Is Seeing Believing?
Digital imaging software could unlock a new realm of visual biomarkers for detecting melanoma
At a Glance
- Only one in 10 excised skin lesions are found to be melanoma
- New automated software allows high-sensitivity, in vivo detection of melanoma using visual biomarkers
- The software is intended as a pre-biopsy visual diagnostic for dermatologists and non-experts alike
- The software will grow and improve as more lesion images and visual biomarkers are added.
Patients with skin anomalies often have only one question on their minds: mole or melanoma? Is this strange-looking freckle a benign dermatological feature or the frightening cancer they’ve heard about on television? Obviously, there are a few ways to tell the difference that even non-dermatologists can use – one of which is size. As a general rule, we know that melanomas tend to be larger than benign nevi... or do they?
Some time ago, a dermatologist colleague mentioned to me that a group of melanomas he and his colleagues were investigating may have actually been smaller, on average, than nevi in the same patients. To test their suspicion, they asked me to create an automated computer vision app to screen the sizes of the lesions. They selected a group of images of “difficult” lesions they had biopsied – nevi that looked highly suspicious, or melanomas that didn’t. These are called “clinically equivocal” lesions. It turned out that they were right; on average, the melanomas were indeed smaller than the nevi in the group of images. Of course, that only holds for this study group, not the general population, and large moles should always be evaluated by a certified dermatologist. But at least in this instance, it seems that some cancers don’t play by the “rules” – and that anomaly piqued my interest.
I wanted to be able to quantify and validate what we were seeing with a sensitivity and specificity analysis, so I took my first step towards creating digital imaging software that uses visual biomarkers to screen for melanoma (1). And there is a real need: at the moment, only 10 percent of excised skin lesions actually turn out to be melanomas (2), which means the vast majority of those patients are undergoing unnecessary biopsies that not only use up medical resources but also leave them prone to infection, scars, or even reduced mobility. Through the eyes of an expert dermatologist, lesions can be classified quite accurately – but current visual diagnostic methods (see “Current Classification Guidelines”) result in a much lower success rate for other professionals. Why? In the United States, there’s one expert dermoscopist per 6.5 million people (see Table 1). No one can see that many patients in a year, so it’s clear that not everyone who should be evaluated by an expert can be; demand far outstrips supply. I wanted to build a better mousetrap – a diagnostic that would be highly sensitive and specific no matter who was using it. Along the way, I created something even more valuable: a software program that not only delivered superior diagnostic accuracy in our study test group*, but has the potential to help the observer by showing what features it is analyzing and why.
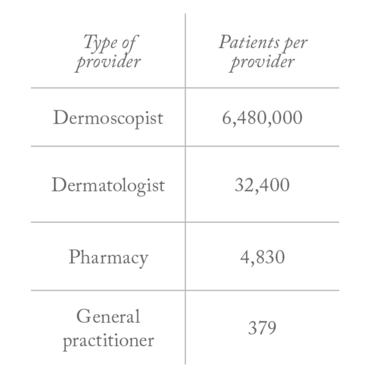
Table 1. Number of patients per type of practitioner (based on US population data).
Current Classification Guidelines
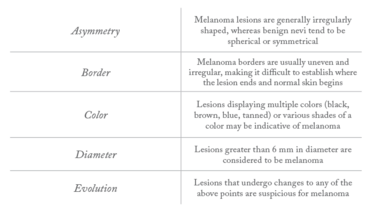
ABDCDE

CASH
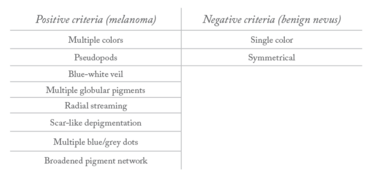
Menzies Method
Finding form
To create a visual biomarker algorithm, you first need to discover initial patterns. My journey began with the acquisition of 120 samples. Next, I sat down, brewed a cup of coffee, and spent 500 hours writing computer code to quantify patterns – fractal arrangements of pigment networks, little islands of the globular pigmented phenotype, and more. I tried to mathematically describe any deviations from “normal” nevus patterns and gave a basic visual estimation using those patterns.
Clearly, no doctor has that kind of time to spend analyzing each sample. Instead, I created a code to find, for example, every pigmentation island and its diameter, so that I could calculate a coefficient of variation (the standard deviation of the island radii divided by the mean island radii). With the help of my colleagues, that coefficient – along with many other visual biomarkers – became the basis for a network of algorithms that generate a quantitative figure (Q-score) to differentiate melanoma from benign nevi. Visual biomarkers including pigmentation patterns and color contrasting with surrounding skin were taken into consideration, but of the 50 visual biomarkers collected, we found that the most significant was the lesion color variation – the number of colors that existed within the lesion.
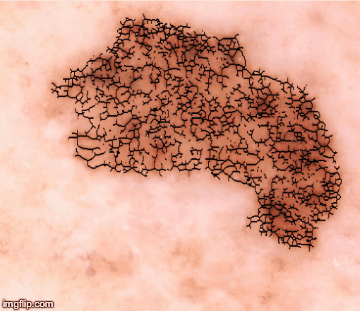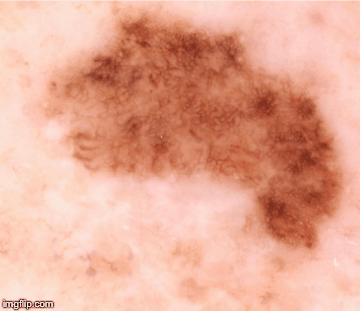
The Q-scores range from zero to one; the higher the number, the more likely a diagnosis of melanoma. The software, used in conjunction with a dermatoscope, reached 98 percent sensitivity and 36 percent specificity in our initial publication; since then, we’ve improved the specificity significantly on that same data set and added additional data sets that confirm the reproducibility of the visual biomarkers as highly discriminant of melanoma versus nevus. Because this approach isn’t based on cellular morphology, it’s not poised to replace pathologists – it’s a guide and a complement to tissue biopsy, not an alternative. What it can do is approach the same level of visual diagnostic ability an expert dermatologist has, with the added benefit of automation so that other medical professionals can use it.
Despite its highly sensitive and specific diagnostic performance, the current iteration of our technique has a few drawbacks. There’s timing, for one. It can take between one and 10 minutes to image a single lesion, depending on its size and complexity – but I think all that’s needed is a few optimization engineers cranking on the code to get the speed down to under 10 seconds for even the toughest lesions. Another concern is the technique’s usefulness in patients with darker skin; our software takes into account color contrasting between the lesion and surrounding dermis, so we need to figure out how to handle different background skin colors. Finally, there are areas of skin that look altogether different – the palms of the hands, the soles of the feet – and the algorithm has yet to be tested on lesions from these areas.

One exciting aspect of ongoing work is the extension of these analytical imaging biomarkers beyond what the human eye (and standard cameras) can see. We’re currently running a multicenter clinical trial to evaluate the melanoma advanced imaging dermatoscope, a hyperspectral camera; preliminary data on about 100 study participants indicates that the imaging biomarkers extend effectively into the ultraviolet and infrared imaging ranges.
The bigger picture
There are many different processes going on in melanoma of which we have only limited understanding from a biological and diagnostic perspective. Our study seems promising, but it’s far from being the standardized technology we need so badly for melanoma detection. Our results are still relatively preliminary, so we need to test the software in wider populations to establish its limitations and reduce the likelihood of significant false negatives. We really need 100 percent sensitivity and at least 50 percent specificity to be ready for prime time. In time, we may even be able to look into connecting these imaging biomarkers more strongly with the underlying biology of melanoma to create an even more powerful version of our tool.
Right now, we’ve taken the first step toward proving the principle that a “fingerprint” of imaging biomarkers can be used like a molecular biomarker. And as we present the software with more examples of known lesions, it will become more familiar with various cancer subtypes – and as a result, its effectiveness will increase. What do we need to take advantage of this technique? A community of people who can help us add as many cases as possible. That’s where you come in.
Do you (or does your colleague) use dermoscopy to guide biopsy of suspicious pigmented lesions? If so, please feel free to contact me (dangareau.net); the next step in our investigations requires collaborators who have a folder of images with a spreadsheet of correlating diagnoses. We can provide a software program to prepare the images for our growing machine learning database. This venture is non-commercial, but academic credit and the satisfaction of contributing to a potentially life-saving research program are assured!
Full-Body Diagnostic Imaging
The field of imaging biomarkers is a relatively new one, but complementary diagnostics like ours are advancing it rapidly. Numerous research groups are now working on total body photography – currently the best way to carry out machine-aided melanoma detection. Rather than generating a static evaluation of one lesion at a time like our system, their method involves taking multiple pictures of the entire body’s surface. The method doesn’t stop there, either; the photographs are updated at regular time points so that diagnosticians can spot the differences – allowing them to detect neoplasms and suspicious changes early on.
It may sound ambitious, but I truly believe that a collaborative team could hone the software’s ability to the point where it creates a valuable visual diagnosis. Whereas other diseases may not be diagnosable without analyzing a plethora of factors, the unique visual biomarkers of melanoma – a disease that starts just a hair’s depth from the skin’s surface – may give us a fighting chance at a one-stop diagnostic tool.
*This technology has not been reviewed by the FDA and should not be used in place of certified dermatologist evaluation.
- DS Gareau et al., “Digital imaging biomarkers feed machine learning for melanoma screening”, Exp Dermatol, [Epub ahead of print] (2016). PMID: 27783441.
- G Salerni et al., “Meta-analysis of digital dermoscopy follow-up of melanocytic skin lesions: a study on behalf of the International Dermoscopy Society”, J Eur Acad Dermatol Venereol, 27, 805–814 (2013). PMID: 23181611.
Dan Gareau is an instructor in clinical investigation at The Rockefeller University, New York, USA.