Studying the brain can be an intimidating prospect, even for neuroscientists. This mysterious organ is composed of a complex network of tens of billions of cells – all transporting electrical and chemical messages over vast distances in milliseconds. Understandably, measuring this activity has not been easy – but, in the last decade, electrophysiology research has become more accessible, thanks to the introduction of microelectrode array (MEA) technology.
MEAs allow neuroscientists to study broad network activity on the lab bench, recording information that was previously out of reach. Historically, scientists who sought to measure the electrical activity of neurons could only look at one or a handful of cells at a time, failing to capture the immense richness of network activity in the brain and beyond. This limited their ability to create complex models of neurological disease in vitro. Furthermore, traditional electrophysiological techniques can take up to a year to master – meaning that most labs cannot readily study neural activity at this level. That’s where MEAs come in; the technology is accessible to most neuroscientists seeking to study the electrical properties of their cells.
Studying neural circuits at the bench
In many ways, neurology lags behind other medical specialties in understanding the nuances of disease and effective treatment. For example, around 65 million people have a form of epilepsy – but there are currently no biomarkers that can predict what treatment is most likely to work for a given individual (1). Rather, physicians must rely on trial and error to find an effective therapeutic. For around one-third of these patients, their prescribed treatment does not work – demonstrating that we still have a lot more to learn about how the brain works in patients with epilepsy (2). Meanwhile, one in 50,000 people develop amyotrophic lateral sclerosis (ALS) but, with no definitive diagnostic criteria or approved treatments, patients face an uphill battle against the disease (3).
To understand and treat diseases such as epilepsy and ALS, scientists need to study neural network activity under various conditions. The standard approach to measuring this activity in vitro is a complex method called whole-cell patch clamp electrophysiology. With this technique, the scientist inserts a micropipette into a neuron in a slice of brain tissue or in culture to measure voltage or current. This technique’s limitations are clear: it’s invasive, because the pipette needs to perforate the cell membrane; cell activity can only be recorded over a period of minutes; and only a handful of manipulations can be reliably tested before the cell dies. Though the method is suitable for understanding the electrical properties of single neurons or ion channels – especially in response to stimulation – and the impact of genetic and pharmacological manipulations on this activity, it cannot measure network activity over long periods. Ultimately, this limits the kinds of questions that a neuroscientist can ask – and answer.
Moving forward with MEA
MEA technology represents a complementary approach to studying the brain in vitro. In essence, an MEA is a grid of tightly spaced microelectrodes embedded in a substrate or culture surface. Traditionally, MEAs have been used to record network activity on the brain’s surface in living animals – a challenging technique to implement. But MEAs can also be embedded into the wells of a multi-well plate, making extracellular recordings of cultured neurons much simpler.
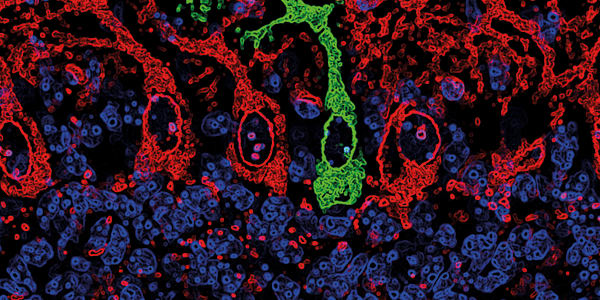
Neurons and other electrically active cells can be cultured over the electrodes, creating a cohesive network (see Figure 1). Researchers can then noninvasively record the electrical activity of this network over weeks and months. The multi-well plate format allows them to grow replicates of the culture and simultaneously test multiple genetic, pharmacological, and environmental manipulations on the network (see Figure 2). Unlike more technically challenging methods, this technique is accessible to any scientist who uses cell cultures. This includes neuroscientists like Evangelos Kiskinis, assistant professor of neurology at the Northwestern University Feinberg School of Medicine, who studies epilepsy and ALS using induced pluripotent stem cells (iPSCs).


Epilepsy in a dish
To study epilepsy effectively, Kiskinis wanted to create a model system that mirrors the disease’s pathophysiology. Previously, scientists have relied on non-human cells or animal models to study the disease – but these fail to capture the patient-specific nuances of epilepsy, relevant diagnostic biomarkers, and druggable targets. Kiskinis solved this problem by developing patient-specific models of epilepsy in vitro using patients’ own stem cells and differentiating them into neurons. By using MEAs to study their electrical patterns and responses to treatment, Kiskinis hoped to identify predictive biomarkers that could help physicians determine the most effective drug for each patient.
In his research, Kiskinis examined KCNQ2-associated epilepsy. In building his model, he aimed to determine whether iPSC-derived neurons harboring mutations in the KCNQ2 gene reflected the characteristic firing pattern seen in children with the disease. Children with KCNQ2-associated epilepsy exhibit a characteristic EEG pattern known as burst-suppression activity – highly synchronized firing (“bursting”) followed by intermittent periods of low activity. To assess this pattern, Kiskinis and his team cultured patients’ neurons on MEAs in a multi-well plate and recorded spontaneous firing over several weeks in culture. They found that these neurons exhibited a burst-suppression firing pattern highly reminiscent of the patient’s EEG (see Figure 3) – effectively recapitulating “epilepsy in a dish” (4). This model system provided a phenotypic platform for assessing potential therapeutic options.

Antiepileptics for ALS
In another branch of his work, Kiskinis used MEAs to identify an antiepileptic drug that might help slow the progression of ALS. First, his team used MEAs to study the nature of iPSC-derived neurons from ALS patients that featured the SOD1A4V mutation (an “ALS in a dish” model) – finding that these neurons were hyperexcitable (5). He used other wells on his plate to block inhibitory neurons, but found no effect, indicating that they were not involved. He also grew neurons with a corrected SOD1 gene and discovered that the cells’ aberrant electrophysiology returned to normal.
The antiepileptic drug ezogabine is known to reduce neuron excitability, but Kiskinis was eager to uncover whether the drug had the same effect in ALS patients. Based on his previous work with MEAs, he was able to advance the drug straight into a phase II clinical trial (6), in which he found that neuronal excitability may be a useful biomarker for ALS drug discovery. Now, he and his team want to investigate whether ezogabine can slow ALS progression.
It doesn’t end there
MEAs have also enabled researchers to predict treatments for inherited erythromelalgia (IEM) – a rare neurological disorder also known as “Burning Man” syndrome. Like epilepsy and ALS, treating IEM is not straightforward – existing painkillers do not relieve the constant, burning pain patients feel in their hands and feet. The disease is caused by a mutation in a sodium channel gene that leads to hyperactive pain-sensing neurons. Yang Yang, assistant professor of medicinal chemistry and molecular pharmacology at Purdue University, has identified over two dozen mutations in this gene, suggesting that a one-size-fits-all approach to treatment would not be suitable.
Through trial and error in the clinic, neurologists have determined that an anticonvulsant drug called carbamazepine reduces symptoms in IEM patients with a specific mutation, Nav1.7-V400M. Yang and his team used MEAs to investigate whether other mutations also respond to the drug (7) and, using their multiplexing capability, they were able to test carbamazepine on pain-sensing neurons that harbored several different mutations, including the Nav1.7-S241T mutation, located adjacent to Nav1.7-V400M.
Because IEM is triggered by warmth, Yang studied the impact of carbamazepine on the relationship between temperature and neuron excitability (see Figure 4). As expected, a temperature-dependent increase in activity was evident in his cultures for each IEM mutation, but the drug reversed this effect for the Nav1.7-S241T mutation. In a subsequent clinical trial, Yang found that patients with this mutation reported reduced levels of pain after taking the drug.

MEAs cannot replace patch clamp electrophysiology – but they empower neuroscientists to ask and answer different questions. They enable all neuroscientists, even those who do not specialize in electrophysiology, to study the intrinsic properties of neural circuits and efficiently monitor their response to manipulation. As evidenced by the research described here, MEAs can help create complex models of neurological disease and facilitate the discovery of new biomarkers and treatments for diseases that have confounded scientists for far too long.
References
- A Ngugi et al., “Estimation of the burden of active and life-time epilepsy: A meta-analytic approach,” Epilepsia, 51, 883 (2010). PMID: 20067507.
- J Couzin-Frankel, “As epilepsy drugs fail nearly one-third of patients, scientists seek root causes of seizures” (2019). Available at: https://bit.ly/3mwYzJ9.
- P Jones, “FYI: Epidemiology of ALS and Suspected Clusters” (2020). Available at: https://bit.ly/2RfXzh3.
- D Simkin et al., “Dyshomeostatic modulation of Ca2+-activated K+ channels in a human neuronal model of KCNQ2 encephalopathy,” eLife, 10, e64434 (2021). PMID: 33544076.
- B Wainger et al., “Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons,” Cell Rep, 7, 1 (2014). PMID: 24703839.
- B Wainger et al., “Effect of ezogabine on cortical and spinal motor neuron excitability in amyotrophic lateral sclerosis,” JAMA Neurol, 78, 186 (2021). PMID: 33226425.
- Y Yang et al., “NaV1.7 as a pharmacogenomic target for pain: Moving toward precision medicine,” Trends Pharmacol Sci, 39, 258 (2018). PMID: 29370938.
- P Geha et al., “Pharmacotherapy for pain in a family with inherited erythromelalgia guided by genomic analysis and functional profiling,” JAMA Neurol, 73, 659 (2016). PMID: 27088781.