
It’s not every day that you have the opportunity to meet a true pioneer of medical technology. But Mark Driscoll – having developed one of the very first next-generation DNA sequencers (NGS) – certainly falls into that category.
After leading the way on research into the human genome for many years, he switched his attention to using NGS technologies to investigate the human microbiome – and Intus Bio was born.

It was after Driscoll had published a paper on using microbiome NGS to improve infection control in a neonatal intensive care unit that he teamed up with biotechnology business leader Paul Denslow. Denslow recognized the life-changing potential of the technology and had the vision to develop it for primary health care use. Joining forces with long-read sequencer developers PacBio, they launched GutID – a microbiome home testing service that delivers actionable insights for patients.
The Pathologist chatted with Denslow and Driscoll to learn more about the GutID technology and its implications for health care.
What is GutID and how does the service work from the patient perspective?
Mark Driscoll: GutID offers an end-to-end microbiome testing service, from sample collection, through genomic analysis, to a comprehensive gut health report.
Patients receive a test kit for collecting a small stool sample, which is then mailed to our testing lab. The sampling method ensures the stability of the samples – they can be stored at room temperature for many months without the bacterial content changing.
Paul Denslow: We extract the bacterial DNA, analyze and interpret it, map it, and send the patient a detailed report of their gut health. It gives scores for indicators like microbial diversity, richness, and evenness, which can be indicators for both gut and overall health conditions. Patients can either use the reports by themselves or work with a health practitioner who can suggest actions to improve the patient’s gut health, which can have a massive impact on overall health.
Who are your customers?
PD: The patients who use the service essentially fall into three categories. First there are the “bio hackers” who are really trying to take control of their own data and their own health outcomes. They already believe in the importance of the gut microbiome to overall health, and want to access the best possible data on that. James Kinross, one of the UK’s top colorectal cancer surgeons and a leading microbiome researcher, is on record saying that our test is the only one he trusts, because it’s complete, accurate, and detailed.
Another group of customers have unexplained gut function issues. Usually they have seen a doctor, been referred to a gastrointestinal specialist, had multiple tests, all have come back negative, and they are still living in pain and discomfort. They want to know why, and what can be done about it.
The third group generally come to us via doctor referrals, when the doctor or the patient thinks that a broader health condition might be linked to gut health, and wants to either rule it in or out.
What are the technologies powering the GutID platform, and how do they work together to achieve strain-level resolution?
MD: The first challenge is cracking open the bacteria in the sample to release the DNA. We’re dealing with millions of different types of bacteria: some fall apart easily, and others require beating with a hammer, so to speak. But when going in heavy to crack the tough bacteria, you don’t want to destroy the DNA in the weaker ones. We had to invent a whole new system for doing that. So the first technology we use is our patented high-throughput cell lysis method.
Next comes the DNA sequencing. We use PacBio long-read sequencers because they give us exactly the right information for identifying the bacteria at strain level. We worked with PacBio to increase the capacity of the sample hoppers to maximize efficiency in our workflow.
The sequencing results are massively data heavy, so we use machine learning to analyze it. Our machine learning tool was developed in house, and performs analysis, assembly, and mapping of the results. It is also trained to recognize cancer biomarkers.
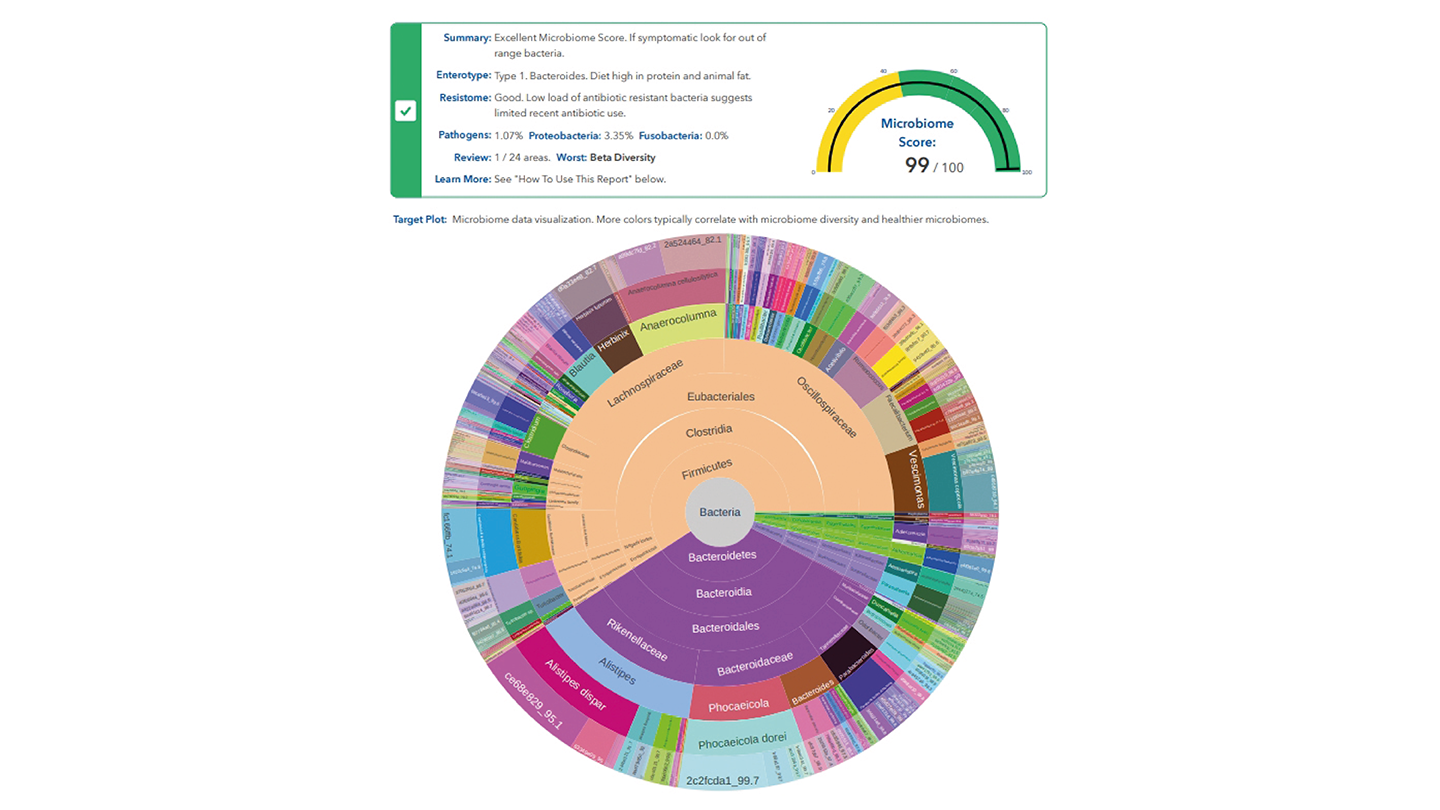
PD: The combination of the highly accurate long reads from the PacBio DNA sequencer and our unique assay allow us to see the entire colony of gut bacteria. We separate each individual strain of bacteria and represent them in the outer ring of our target plot data visualization. From there, we work inwards, mapping species and genus, all the way to phylum and domain.
This strain or outer ring led approach is what allows us to generate data for the entire microbiome – something other approaches can’t do in a practical way. Our machine learning tools then use the data to generate GutID reports, which highlight issues, make recommendations, and contain accurate microbiome health metrics, like evenness and diversity – a true first for the industry.
How do you respond to the criticism that microbiome tests fall short on scientific accuracy?
PD: There are a lot of myths out there that the microbiome is always changing, and is therefore not a reliable source of information. But our research shows that the microbiome is incredibly stable. We have tested samples from the same individuals over time and our assay returns virtually the same result month after month.
These results also speak to the incredible stability of our assay. We have seen repeatability testing with other assays that show very different results from the same sample. If we run two tests from one sample on our system, the results are essentially identical.
Ours is a very data-driven company. Scientists should be celebrating the insights our assays generate because they are backed up by solid data. Our technology is transforming an area of health care that was previously very ambiguous and gray into one that can be quantified and monitored.
Could you elaborate on how the GutID test results are translated into clinical or lifestyle interventions for the patient?
MD: I can give you an example where I was the test subject! As inventor of the test, I wanted to take the test and prove that my microbiome is excellent. However, the test results showed straight away that it’s not so great.
In the interest of science I retested my microbiome every month for 14 months, without making any lifestyle changes, and – sure enough – the results kept coming back the same. This demonstrates what Paul was saying: if you just live your life, the microbiome is incredibly stable.
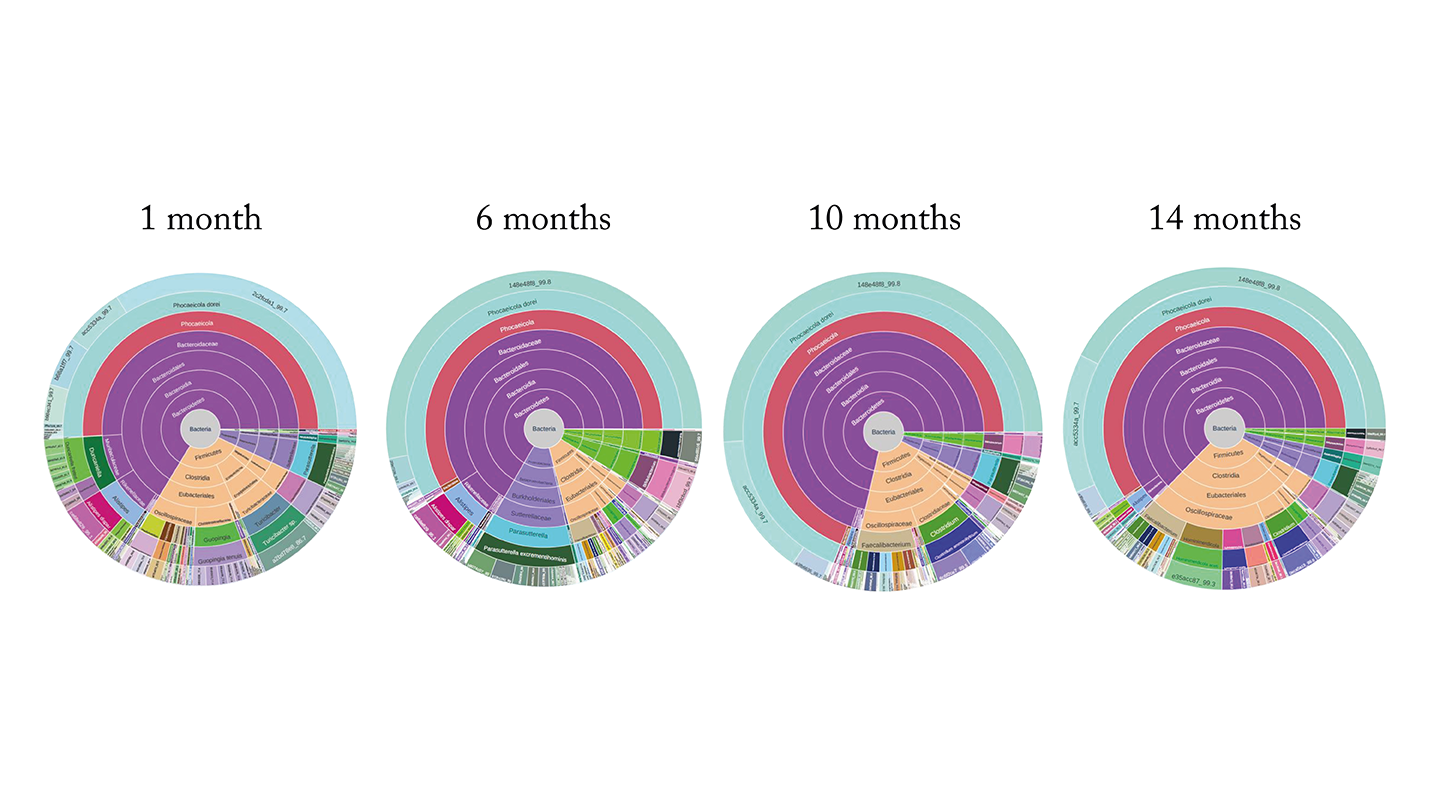
For years I had experienced gut pain after eating. And I’d never really addressed it – it was just part of my life. When I showed my microbiome map to Intus Bio’s nutritionist, Elena Panzeri, she started asking me about my diet and lifestyle. I explained that I’m healthy; I run, I lift, I eat well. She said, “But you’re not eating any vegetables.” I said, “I am!” Well, it turns out that peanuts are not vegetables.
She recommended taking a very specific supplement to make up for the deficits in my diet. I thought, “Given the stability of my microbiome for the past 14 months, this is never going to work.” But, sure enough, when I ran the GutID test just a few weeks later, the change in my microbiome was very noticeable. What’s more – I felt better. The post-meal cramping that I had experienced for years totally went away. I was able to walk the dog and help with the dishes after meals, instead of lying on the couch clutching my stomach.
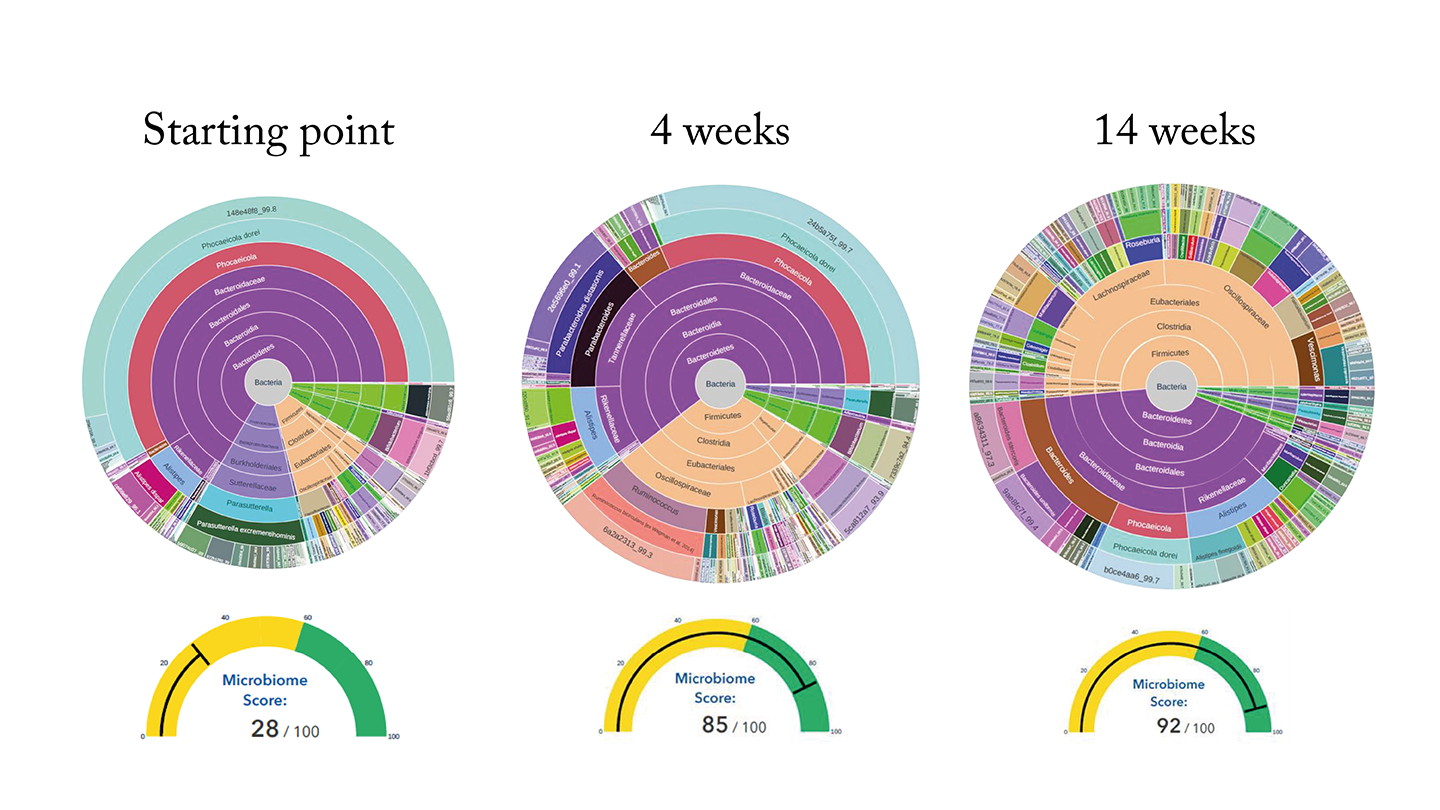
The GutID test reports do make some general recommendations for patients. But my experience demonstrates the value of talking to a qualified professional who knows your story in order to hear a personalized recommendation.
PD: We believe in presenting microbiome data in an actionable manner. Clinicians don’t want to spend hours wading through data – they want to see the headlines at a glance, on the first page of the report. Is there a problem and does it need further investigation? Our reports show target plots and microbiome scores on page one, which indicate from the offset whether an intervention might be required. Subsequent pages give more detailed information on the biodiversity, beneficial bacteria, disease-related bacteria, species related to digestive health, and so on, which can inform the nature of those interventions.
The other way microbiome testing helps is in proving whether the interventions are working or not. A patient’s doctor or dietitian might recommend they take a probiotic. But there are so many different ones on the market – how does the patient know which one to pick, and whether it’s making a difference? Mark’s example of “before and after testing” shows that our test provides the answers to those sorts of questions.
How do you see strain-level microbiome analysis impacting broader areas of healthcare?
PD: The first group of clients we had for GutID in the UK were all being treated for mental health conditions. They wanted to understand the impact of gut health on those conditions. A lot of patients in the mental health space have gut function issues. We can show them the link between those issues and their microbiome, and present something that can be measured and improved – gamified, in a way. That can be very powerful.
We are now doing some research with The Delamere Clinic in the UK with patients with alcohol use disorder that is showing some very strong signals.
MD: We’re also working with some leading cancer centers in the States, looking at microbiome testing for diagnosis of early-stage pancreatic cancer. Because the pancreas dumps into the fecal space, we can detect the bacterial biomarkers in the poop, and then we can give an early warning for disease. And it’s a non-invasive test.
If approved, we could build it into our microbiome analysis software so that all of our customers are screened for pancreatic cancer as standard.
PD: We have this incredibly powerful platform, and it's very, sample hungry. If we can access any kind of samples of specific patient cohorts – be it spit, stool, swabs, even tissue – we can run them on our analyzer. And the results will very clearly show whether or not there’s a microbiome correlation with the condition in question.
So the thing that is holding our research back is not the technology, it’s the samples to train and build the data to run these applications. In that respect, here is a call to action for readers of The Pathologist: if you are looking to develop an understanding of the link between the human microbiome and a specific condition, and you have samples from a cohort, please get in touch.
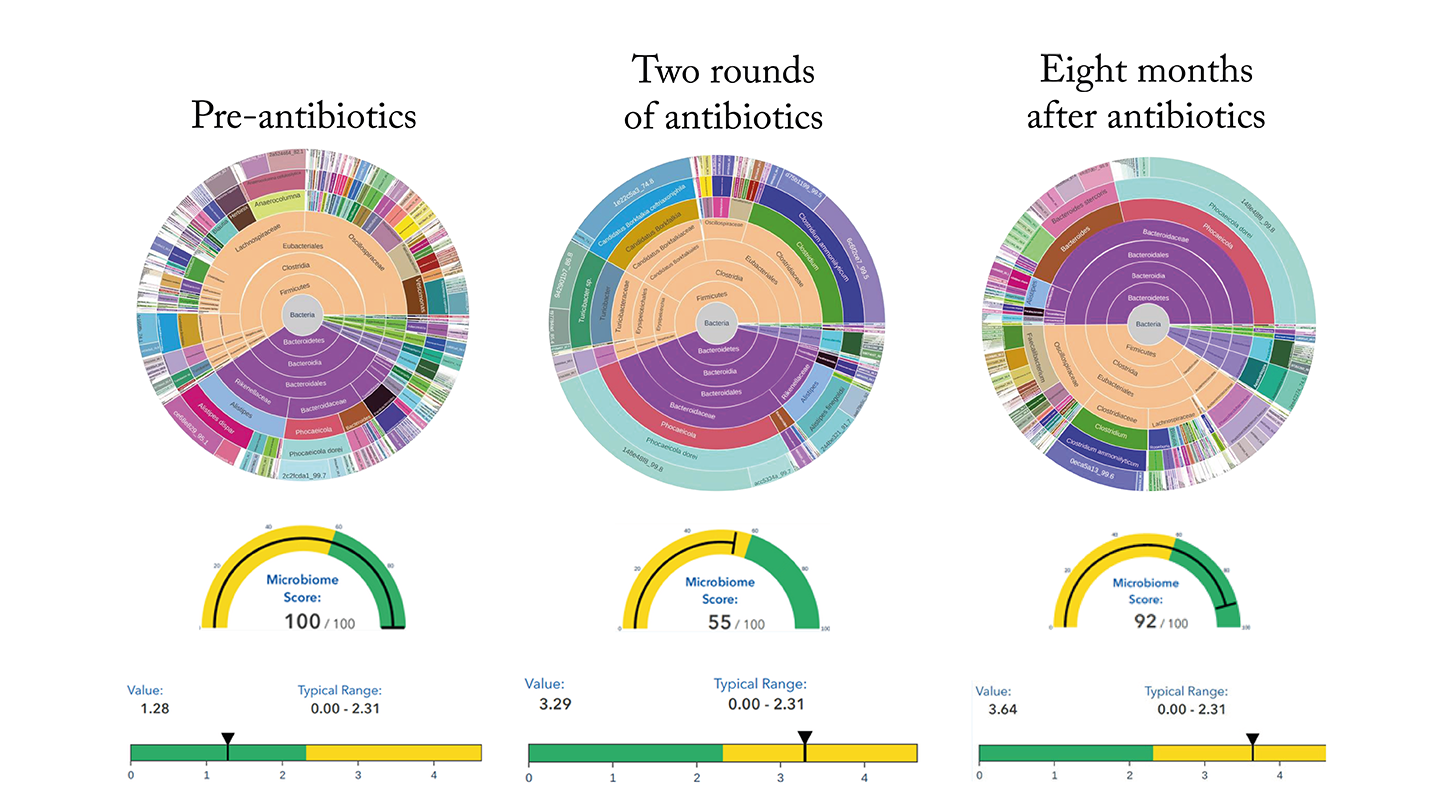
Case study 1 – antibiotics
This patient had an amazingly healthy microbiome with an overall score of 99. Her resistome score – the number and types of bacteria that are antibiotic resistant was also deep into the green end of the scale, as shown on the left.
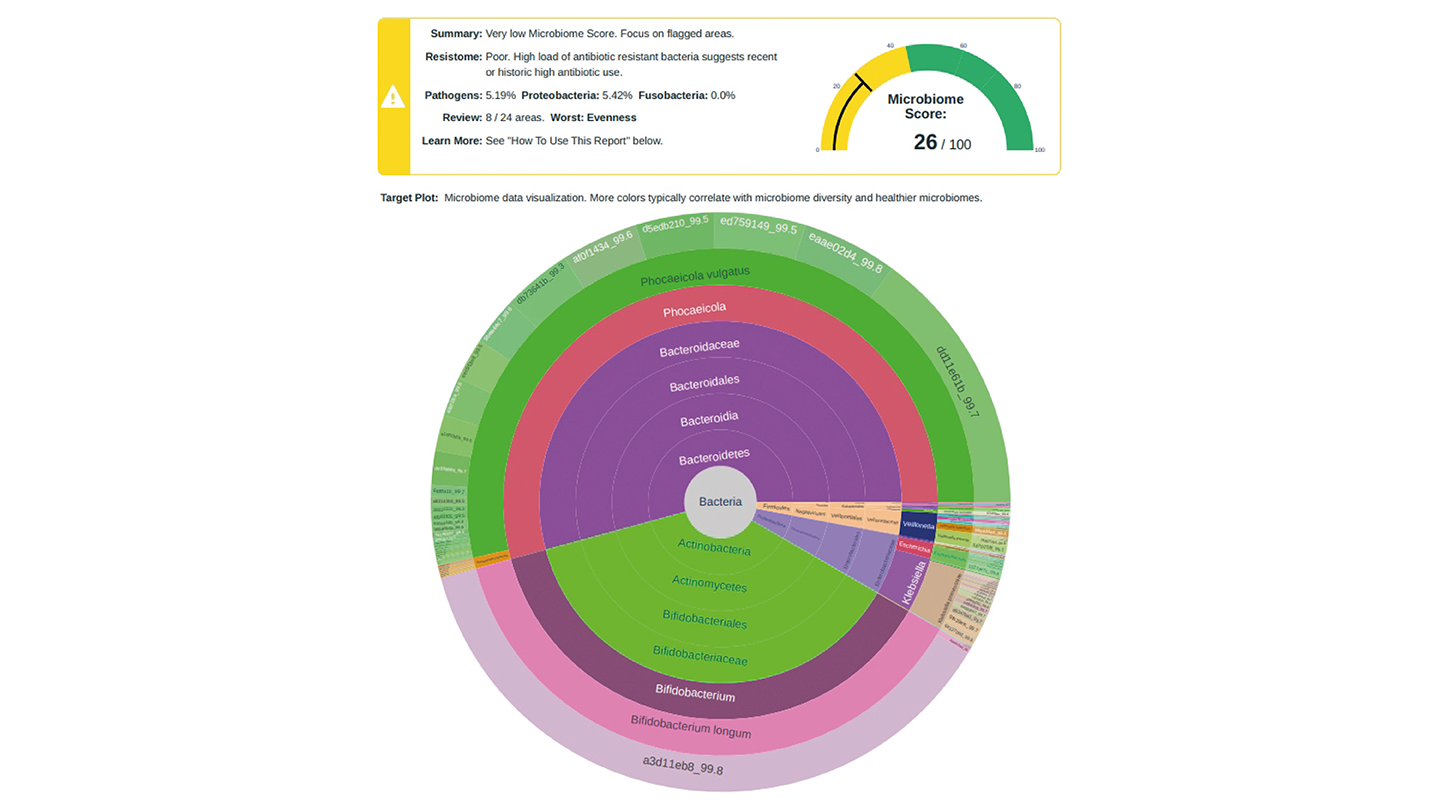
The middle image shows test results from the same patient after two courses of antibiotics for an infection. Whilst the treatment was essential for her wellbeing, it had a profound effect on her microbiome.
The results showed that action was required to restore the patient’s microbiome. She was advised to take a dietary supplement, and her microbiome started to recover, as shown on the right.
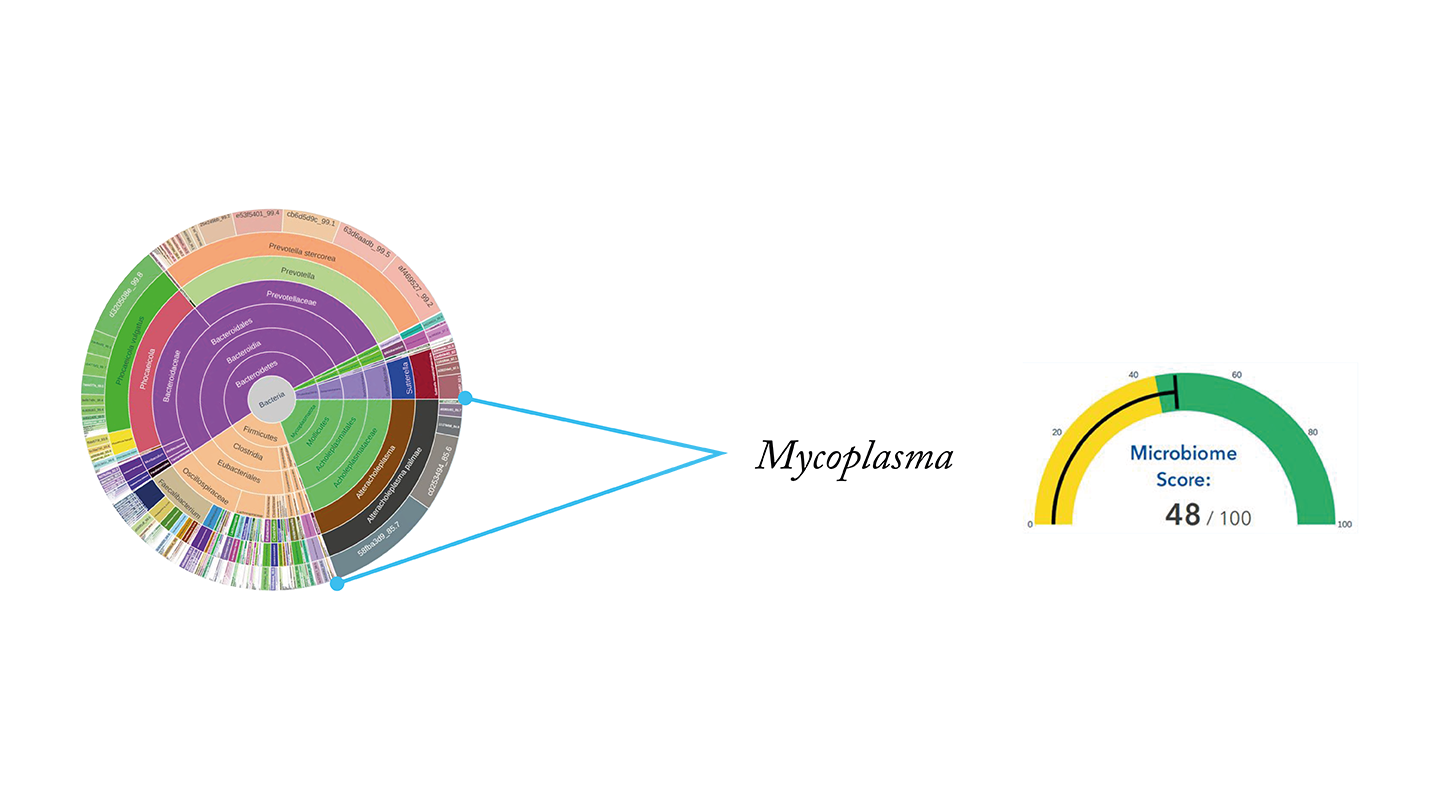
Case study 2 – chronic diarrhea
This patient suffered from chronic diarrhea for around 2 months, for which her doctors could offer no explanation.
Her GutID test results revealed a colony of pathogenic Mycoplasma, which explained the symptoms but had not been picked up by any other tests.
After taking a targeted supplement and probiotics for a few days, the patient reported relief from the symptoms.
Case study 3 – anxiety
This professional athlete suspected that anxiety was affecting his performance. His GutID test results identified an overgrowth of Alistipes – a bacteria linked to anxiety.
His dietitian recommended some dietary changes to drive down the Alistipes population and enhance the abundance of Firmicute. He also introduced specific foods to his diet to increase probiotic Akkermansia and Lactobacillus.
After making the dietary changes, the patient reported reduced symptoms of anxiety, and an improvement in his performance and rankings.
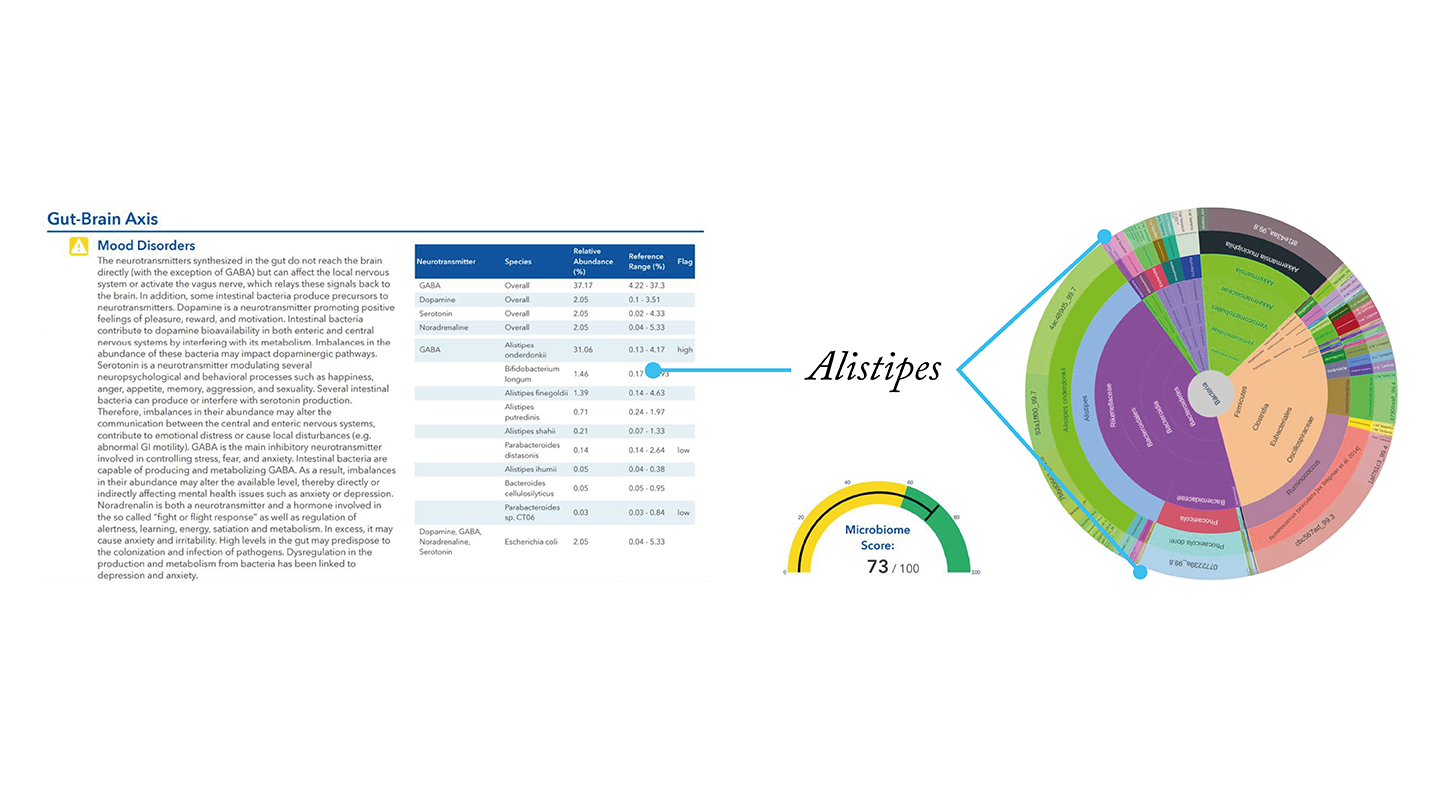
Image Credit: Intus Bio




