Could DNA methylation analysis provide a new method for early detection of non-small cell lung cancer? A recent study, published in Genes and Diseases, explores the possibility.
The researchers, based in China, analyzed tissue and plasma samples from patients with lung cancer and benign conditions. Using bisulfite sequencing, they identified differentially methylated markers associated with lung cancer. The team then developed diagnostic prediction models based on these markers and evaluated their effectiveness by measuring sensitivity, specificity, and overall diagnostic accuracy.
The study identified 276 significantly differentially methylated markers in lung cancer tissue. A diagnostic model using six of these markers achieved a sensitivity of 90 percent and specificity of 97 percent in the training cohort, with similarly high performance in the validation cohort.
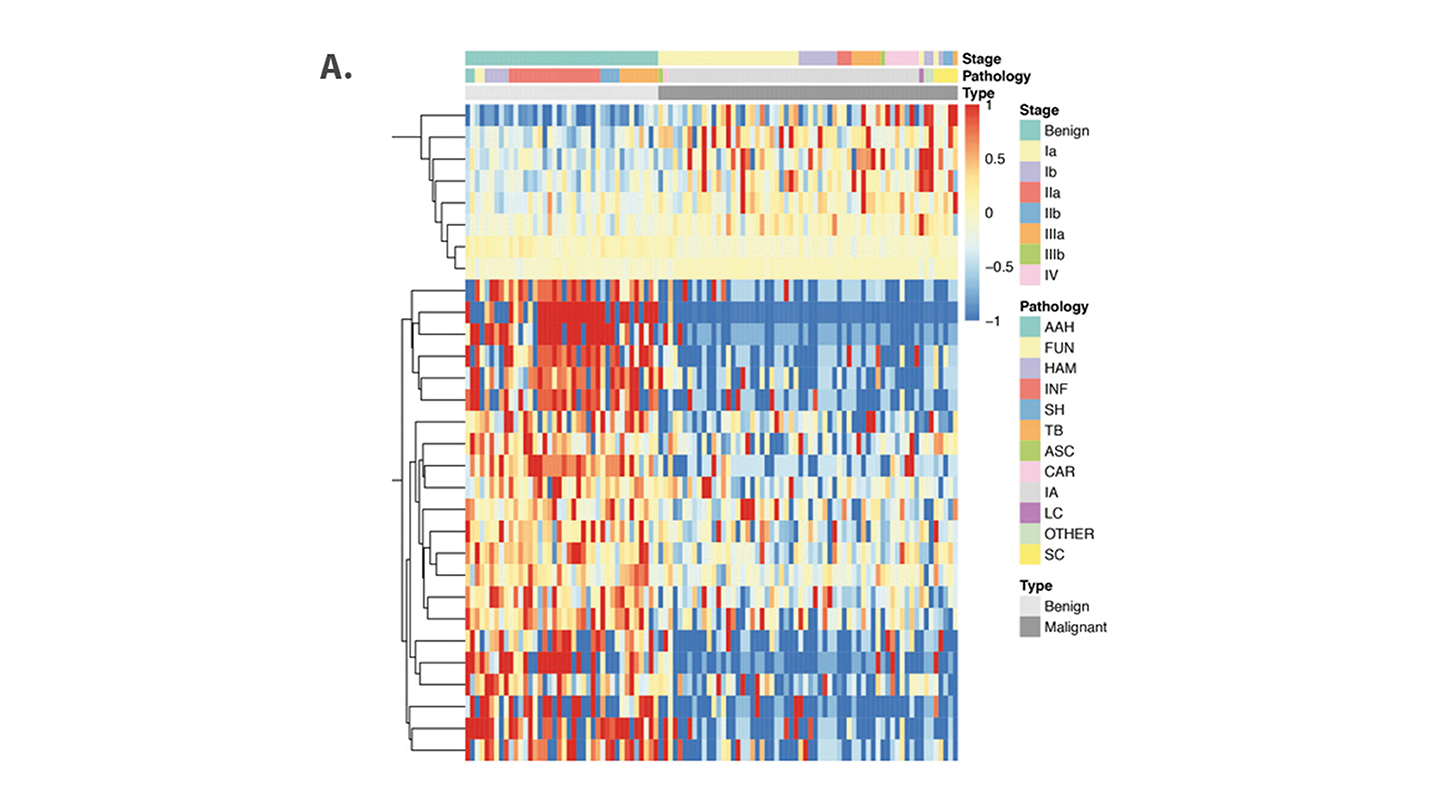
In plasma samples, a separate model using nine markers showed strong diagnostic potential, with a sensitivity of 98 percent and specificity of 100 percent in the training phase, though performance dropped in the validation cohort (sensitivity 81 percent, specificity 59 percent).
Compared with traditional biomarkers like carcinoembryonic antigen (CEA) and neuron-specific enolase (NSE), the methylation-based models demonstrated superior accuracy. Additionally, the plasma-based test offers a non-invasive alternative to tissue biopsy, making it a practical option for routine screening and monitoring.
Though DNA methylation analysis shows diagnostic value, further validation with larger patient cohorts is needed. Nevertheless, noted the researchers, “By integrating multi-modal information provided by computed tomography images, ctDNA mutations, and ctDNA methylation patterns, both the sensitivity and specificity of early lung cancer diagnosis could be dramatically improved.”




