
Researchers from the UK have recently found a way to distinguish epigenetic changes in the cells of patients with chronic fatigue syndrome (CFS), also known as Myalgic Encephalomyelitis (MS). The study outlines the potential for the first blood test for the debilitating condition. To learn more, we spoke with Alexandre Akoulitchev, Chief Science Officer at Oxford BioDynamics and one of the researchers involved in the study.
What prompted you to explore a blood-based diagnostic test for ME/CFS, and how does this address current challenges in diagnosis?
We were approached by a team of clinical experts in ME/CFS, led by Dmitri Pshezhetskiy, who were familiar with our work in clinical biomarker development. Our group has already translated two high-accuracy tests into clinical practice: one for prostate cancer detection (PSE) (1,2) and another for predicting patient response to the major class of immune checkpoint inhibitors – PD-1/PD-L1 antagonists such as Keytruda – across more than 14 indications (CiRT) (3,4).
Our EpiSwitch platform uses a relatively new biomarker modality: 3D genomic regulatory architecture (5). This approach captures disease-specific network dysregulation at a systemic level using whole-blood samples. EpiSwitch biomarkers have demonstrated robust performance in several complex conditions, including ALS and rheumatoid arthritis.
Because of this track record, our clinical colleagues asked whether we could help identify a diagnostic signature for the highly challenging condition of ME/CFS. With more than 400,000 suspected patients in the UK alone, there is an urgent need for a rapid, reliable, and definitive diagnostic test for this debilitating disease.
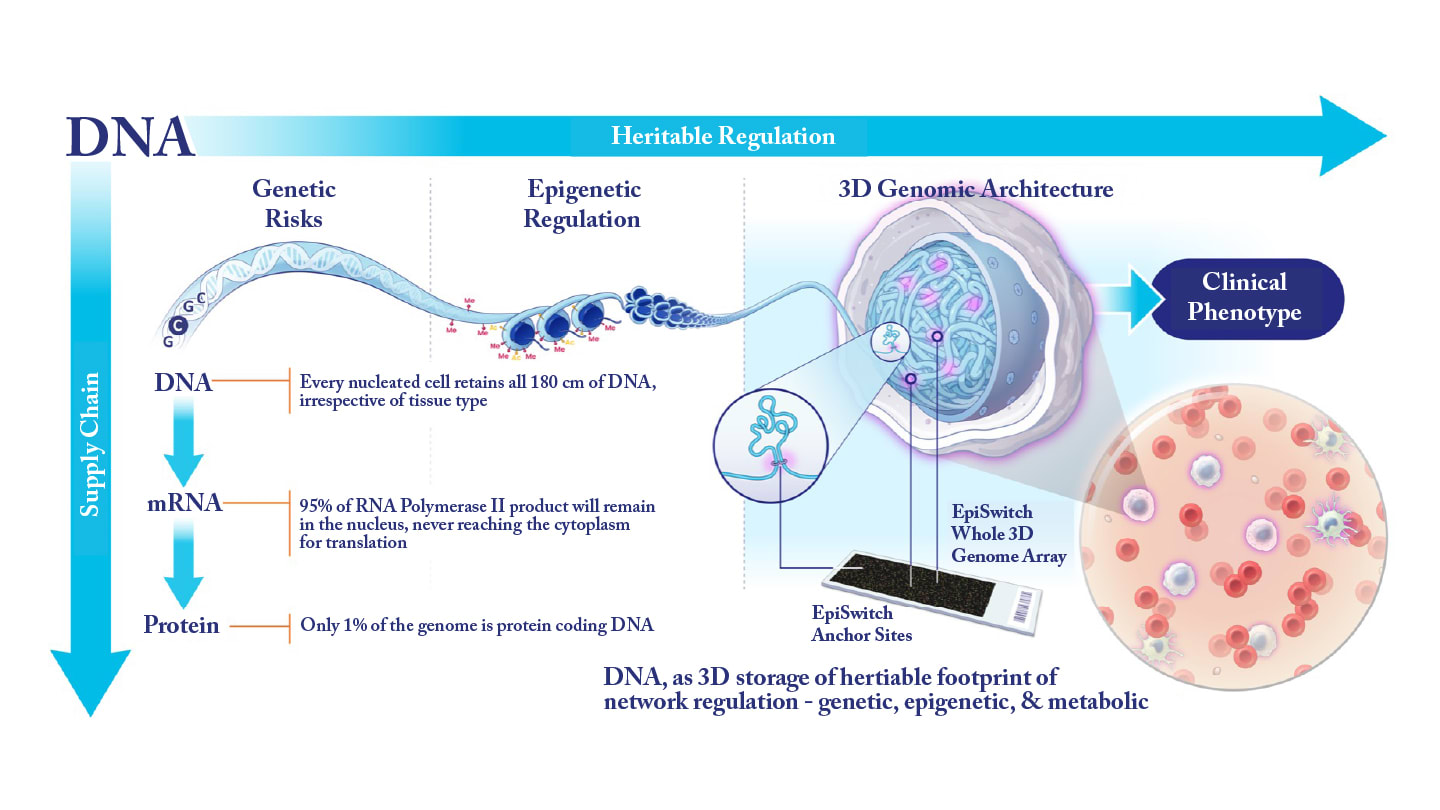
Could you briefly explain the EpiSwitch platform and how 3D genomic profiling differs from traditional genetic or epigenetic biomarker approaches?
Every living cell in the body carries the full complement of heritable information, far beyond the linear genetic code shared by all cells across tissue types. Much of this additional information is governed by epigenetic mechanisms – such as DNA methylation and histone modifications – which influence how DNA is packaged within nucleosomes.
Another crucial dimension of genomic regulation is the way 1.8 meters of DNA are folded and organized inside a nucleus only about 10 microns in diameter. This 3D architecture is highly regulated, and daughter cells reproduce the same structural configuration as the parent cell during division. The long-range interactions formed during this folding process determine which genomic regions are accessible to the transcriptional machinery at the appropriate time and location. These interactions coordinate and juxtapose distant regions of the genome to enable precise regulatory control.
In effect, the 3D architecture of the genome serves as a biophysical “footprint” of a cell’s epigenetic and metabolic state. It reflects intercellular variation – from tissue-specific identity to pathological dysregulation within individual cells. Because cells throughout the body engage in epigenetic crosstalk through metabolic cues, non-coding RNAs, exosomes, and other molecular signals, it is possible to detect stable, condition-specific differences within the 3D genomic landscape that correlate strongly with primary sites of disease.
Cells of the innate and adaptive immune systems, which circulate broadly and sample signals from every tissue, are particularly sensitive to these 3D genomic imprints. As a result, they provide a consistent and informative liquid-biopsy source for biomarker detection. A more comprehensive explanation of this concept is available in one of our recent perspective articles on the technology (5).
Can you tell us about your methodology in running this test?
A standard whole-blood sample collected in tubes containing sodium EDTA or sodium citrate serves as the input material for the EpiSwitch platform. These samples remain stable at ambient temperature for up to 28 days, including during shipping and storage, and can also be frozen long term without degrading test performance. This level of stability is a significant advantage of the technology. The long-range chromosomal interactions – our chromosome conformation signatures – remain reliably detectable as present or absent long after collection.
Once the sample is received, it undergoes processing in which its epigenetic analytes – essentially long-range 3D genomic interactions – are converted into sequence-based readouts within five to six hours. Using our bespoke array to profile the entire 3D genomic landscape, we evaluate the top 200 validated biomarkers. Together, these markers form a tightly connected network of biologically related deregulation – a characteristic molecular footprint of the disease (6).
Given the complexity and overlap of clinical presentations in ME/CFS and related conditions, this approach enables us to distinguish ME/CFS from COVID-19, rheumatoid arthritis, lupus, mast cell activation disorders, fibromyalgia, and PTSD with a high degree of confidence.
What level of accuracy and reproducibility did the biomarker panel achieve in the validation phase?
Having already translated tests such as PSE and CiRT into clinical practice, we recognize the need for extended validation and real-world clinical utility studies before the ME/CFS test can be made commercially available. The high performance observed in this initial study did not surprise us; it reflects a key advantage of using 3D genomic architecture biomarkers. These biomarkers behave as stable, binary signals with a high signal-to-noise ratio, making them far more practical in terms of required sample sizes – based on power calculations and effect sizes – than rare genetic mutations or highly variable continuous readouts from transcriptomic or proteomic assays.
As molecular biomarkers, 3D genomic signatures translate more reliably into clinical outcomes. For context, our PSE test has demonstrated 94 percent accuracy, a result that has held up across several thousand patients and in a recently published utility study (1,2).
Could this biomarker panel be used to monitor disease progression or treatment response?
Our technology has already demonstrated usefulness beyond diagnostic detection alone. Prognostic applications, residual disease monitoring, and treatment-response assessment have all been successfully explored using whole 3D genome profiling. This approach effectively captures an individual’s epigenetic landscape, which influences both disease severity and the biological conditions that determine how a patient may respond to a given therapeutic agent.
Developing such tests requires an unbiased discovery process similar to the one used for the ME/CFS panel, even though some individual markers may appear across multiple disease-specific footprint signatures.
How might the test influence clinical decision-making?
Our understanding is that it can currently take a year or more for patients to receive a diagnosis of ME/CFS. In contrast, an EpiSwitch test could provide a clear result within one to two weeks. This has the potential to reduce patient anxiety, shorten the diagnostic journey, and help clinicians identify an actionable underlying condition more quickly.
Accurate diagnosis is also essential for patient recruitment and therapeutic development. There may be distinct biological clusters within the ME/CFS population, some of which could be more responsive to specific treatments. A recent report describing the successful use of low-dose rapamycin to alleviate ME/CFS symptoms is one example that highlights the importance of having a well-defined diagnostic foundation (7).
What are the next steps for this research?
There is strong interest in applying our technology to additional complex conditions. However, we are equally committed to the path we know well – bringing the ME/CFS test into routine clinical practice and making it accessible to the public.
References
- D Pchejetski et al., Cancers (Basel), 15, 3 (2023). PMID: 36765779.
- J Berghausen et al., Cancers (Basel), 17, 13 (2025). PMID: 40647492.
- E Hunter et al., Cancers (Basel), 15, 10 (2023). PMID: 37345033.
- J Abdo et al., Cancers (Basel), 17, 17 (2025). PMID: 40940997.
- J Mellor et al., Cancers (Basel), 17, 13 (2025). PMID: 40647485.
- E Hunter et al., J Transl Med, 23, 1 (2025). PMID: 41057909.
- BT Ruan et al., Res Sq (2025). PMID: 40502741.




