The field of pathology is rapidly moving toward a digitally enabled future where all pathologists access, view, diagnose and manage cases using whole slide images (WSIs) and artificial intelligence (AI) algorithms. As this transformation progresses, it will continue to be critical for pathologists, laboratory professionals, and other key decision makers to become fluent in the evolving language of AI and digital pathology (DP); after all, we will be charged with making informed decisions about which specific digital and AI pathology solutions best meet the needs of our practice environment.
Achieving this fluency will not require the knowledge to code AI applications but will require the acquisition of foundational levels of knowledge in the areas of DP infrastructure as well as underlying AI, machine learning, and neural network approaches. The focus should be on understanding the strengths and weaknesses, training requirements, and level of supervision associated with each approach. The depth of knowledge should be similar to what pathologists and lab professionals already know about other testing methodologies in routine use, such as immunohistochemistry (IHC), next-generation sequencing (NGS), and polymerase chain reaction (PCR). The goal should be a level of familiarity and comfort in critically assessing different AI solution options and matching specific pieces of digital pathology software, AI algorithms, and other tools to current and future needs in their laboratory environment.
Here, I present my beginners’ guide to AI fluency.
Digital and AI pathology infrastructure
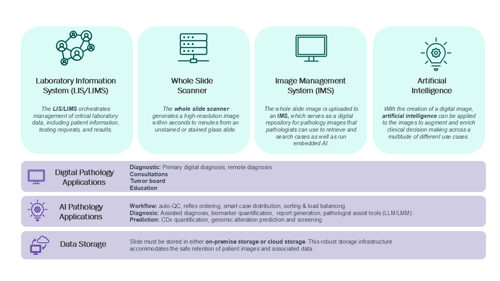
DP refers to the scanning of histopathology glass slides and viewing of digital WSIs. The core components of a DP infrastructure include a digital slide scanner and either a basic digital image viewer or full image management system (IMS; see Figure 1). Digital slide scanners are rapidly evolving, with the latest generation of on-market hardware approaching feature parity from a resolution, throughput, and overall image quality perspective. Ongoing advancements will surely continue, especially in the areas of z-plane processing (which is necessary for cytology and other depth of focus applications) and support for fluorescence imaging.
Once glass slides have been digitized into WSIs, downstream viewing and management requires either an image viewer or full-featured IMS. Most DP scanners are sold with image viewers that enable basic digital workflows, such as primary digital diagnosis, remote viewing, consultations, and education. An IMS goes beyond an image viewer to provide a comprehensive platform for histopathology case distribution, prioritization, management, organization, searching, and consultation. Most IMSs have the ability to interface with the lab information system (LIS) in a bi-directional manner to allow pulling of case and patient metadata into the IMS and pushing of DP and AI data back to the LIS and pathology report.
The IMS also enables the use of AI pathology applications, either natively/directly integrated or via so-called contextual or “pop-out” integrations. It is important to understand the difference between these two scenarios; they may have significant implications for i) real-world ease of use, and ii) the flow of complete data from AI algorithms to the IMS and then to the LIS and pathology report. IMSs with natively integrated AI applications allow immediate and direct use of AI tools within the same diagnostic viewing environment. Whereas, without direct integration, AI applications rely on launching a separate application window where the AI is visualized. From a data flow perspective, IMSs with natively integrated AI solutions by design have the required fields to accept data from the AI application for delivery to the IMS for reporting. A potential limitation with non-native AI algorithm integrations into an independent IMS is a mismatch in either functionality (for example, the AI requires overlay capability not present in IMS) or data field quantity, leading to potential limitations in reporting. Additionally, the creation of AI-IMS application programming interfaces (APIs) may be required, at additional cost and timeline impact, to enable full functionality of a non-native AI application with an IMS.
AI and machine learning methods
AI is a broad field of many methods focused on the use of machines to replicate the intelligence of humans:
Machine learning (ML) is a subset of AI that specifically uses algorithms and statistical constructs to learn from data and improve performance on specific tasks.
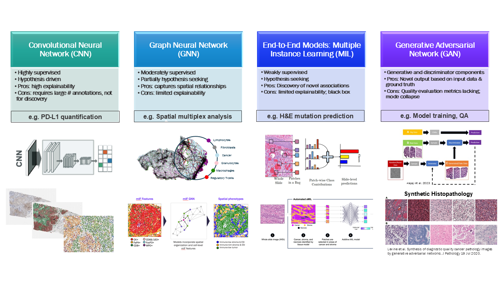
Deep learning (DL) is a subset of ML that uses deep neural networks to process and analyze vast amounts of data to identify patterns and make predictions. Several specific types of deep neural networks have been used to create histopathology-specific AI applications that have the potential to improve the efficiency, turn-around time, reproducibility, and accuracy of pathologist and histotechnician workflows (see Figure 2).
Let’s dig a little deeper into AI model types and their applications:
Convolutional neural networks (CNNs) are supervised ML models trained on large amounts of labeled data that can identify the presence of trained structures in previously unseen data sets. CNNs are hypothesis-driven, meaning one needs to know what the model should be identifying. By design, they will not discover any new associations within the data set or identify structures not present in the training data. An example CNN application in pathology is PD-L1 scoring. Several CNN-based algorithms have been created to identify PD-L1 positive and negative tumor and/or immune cells and output the relevant PD-L1 scoring metrics. They are trained with large numbers of human pathologist annotations (for example, tumor cells positive for PD-L1). However, these models will not discover a new way to predict responders to immune oncology (IO) drugs. Another common CNN algorithm application is tumor detection for quality control (QC), NGS sufficiency, and case prioritization purposes.
Graph neural networks (GNNs) exist between CNN- and MIL-based models in that they are moderately supervised, relying on some identified features but also have the ability to discover new associations in the data set. This approach is based on the creation of graphs, which consist of “nodes” connected by “edges”. In histopathology, the nodes are typically morphologically identified cell types, such as lymphocytes, tumor cells, and so on, but can also be cells identified by IHC expression. The edges can be any relationship between cells but are typically the distance between cells. Multiple graphs, representing different spatial relationship patterns, can be examined for correlations with an endpoint or characteristic of interest, such as drug response, survival, or presence of a molecular biomarker. Example applications well suited to a GNN approach include analysis of multi-analyte multiplexed biomarkers (for example, IHC multiplex) and any biomarker based on spatial location or proximity of cells and architectural features.
Multiple instance learning (MIL) is a weakly supervised, hypothesis-seeking methodology that does not use labeled data but instead can discover patterns in a data set that correlate to an endpoint or characteristic of interest. A pathology example is molecular biomarker prediction where the model is trained to identify cellular and/or tissue-level morphologic patterns in the hematoxylin and eosin (H&E) slide that correlate with a specific gene alteration (for example, a point mutation). MIL has also been used to directly predict drug response from H&E. Advantages of this method include the ability to discover novel associations and the lack of highly annotated data requirements. The main disadvantage is that end-to-end methods like MIL tend to have a low level of “explainability,” meaning they are more “black box” in nature, with the user potentially not having the ability to visually confirm the presence of the cell/tissue features driving the correlation.
Generative adversarial networks (GANs) Generative AI methods create novel output data based on input data and a ground truth. In a GAN model, there are actually two models, one generating novel output and a separate discriminator model comparing the output of the first model with the ground truth. In histopathology, GAN models can be used to transform the WSI output of one scanner model to visually match the output of another scanner model, which can be valuable to cross-train an algorithm to be generalizable to multiple scanner types. Some groups have used GAN technology to create synthetic histopathology images, which may have utility in research, training, and studies (1).
Continuous learning and the future
The examples covered in this brief article are not meant to represent the full spectrum of available ML methods relevant to histopathology today and, of course, cannot anticipate the new and even more sophisticated methods that will certainly be coming in the future. Pathologists and other lab professionals should take advantage of the abundant sources of information online and in the literature to establish and build their DP/AI pathology fluency, one layer at a time. Challenge yourself to learn more about topics not covered here, such as self-supervised learning (SSL), zero-shot learning (ZSL), transformers, foundation models, and visual language models (VLMs).
VLMs represent the cutting edge of this field and offer an indication of where it will go in the future. Pathology VLMs have been trained on vast numbers of histopathology image–text pairs to create powerful pathology-specific chat bots (2). The vision is that these generative AI tools will serve as pathologist assistants during the diagnostic process; for example, to create a differential diagnosis and refine it with recommended additional IHC and molecular testing. In addition, these tools could assist the pathologist in the creation of the pathology report, including drafting the microscopic description, synoptic diagnosis, and any relevant clinical decision support information. Importantly, pathologists should remain in control and be the final decision maker, choosing when and how to use VLMs – or any other AI tool.
Embracing the change
Anatomic pathology has a long history of adopting novel technologies that advance the field and improve our ability to characterize tissue samples and generate information that is valuable to patient care. These include the microscope itself, H&E dye technology, special stains, IHC, PCR, and NGS. Digital and AI pathology are powerful new tools that complement the existing methods pathologists currently use in their daily work. Just as there was a learning curve for the pathology field in the adoption of past technologies, so there will be for DP and AI. Rather than being fearful of (or intimidated by) DP and AI, pathologists and lab professionals should take this opportunity to fully lean in and embrace these techniques; they have the potential to address unmet medical needs across disease areas in ways not previously possible.
As these technologies are implemented, pathologists will certainly need to work cross-functionally within their organizations (especially with information technology and informatics colleagues), but they should take the lead in these discussions and not delegate their decision making authority to others because they feel uncomfortable with the underlying methods. Becoming fluent in the language of DP and AI will enable pathologists to be leaders in this exciting and rapidly advancing field that undoubtedly will bring novel value to pathology departments as well as the clinicians and patients they serve.
Credit: Images for collage sourced from Pexels.com
References
- AB Levine et al. “Synthesis of diagnostic quality cancer pathology images by generative adversarial networks”. J Pathology, 252, 2 (2020). PMID: 32686118
- MY Lu et al. “A Foundational Multimodal Vision Language AI Assistant for Human Pathology”. [Preprint] (2023).




