Advancements in precision medicine are driving the need for comprehensive genomic profiling (CGP). In short, CGP facilitates the simultaneous analysis of a broad range of biomarkers in one test to maximize insights into the underlying oncogenic drivers of cancer. Both common and rare alterations can be assessed in a timely manner, while minimizing the risk of tissue exhaustion associated with sequential testing. Additionally, complex genomic signatures or characteristic patterns of somatic mutations in cancer genomes can be assessed, reflecting the underlying mutational processes of the cancer.
As we gain a deeper understanding of the molecular mechanisms of tumor biology, CGP is critical to help drive insights into advancing the future of personalized medicine.
Research needs assessment
Several questions arise when evaluating CGP assays. What are the advantages of amplicon- versus hybrid-capture-based next-generation sequencing (NGS) methods? Should we choose in-house CGP or send-out services? When making these decisions, it is useful to consider the lab’s individual situation and priorities.
If your lab handles many cytological research samples where tissue is limited, amplicon-based CGP assays may be more appropriate – these have a high success rate of ~94 percent, as opposed to hybrid-capture based methods where quantity not sufficient (QNS) rates up to 30 percent have been reported (1).
If timely results are critical for important insights and decisions, bringing CGP in-house not only greatly reduces the turnaround time – to days instead of weeks – but also allows more control over the sample and preanalytical parameters.
New-to-NGS users may value a high level of automation to reduce labor-intensive steps, as well as integrated bioinformatics to simplify data interpretation without the need for specialized bioinformaticians.
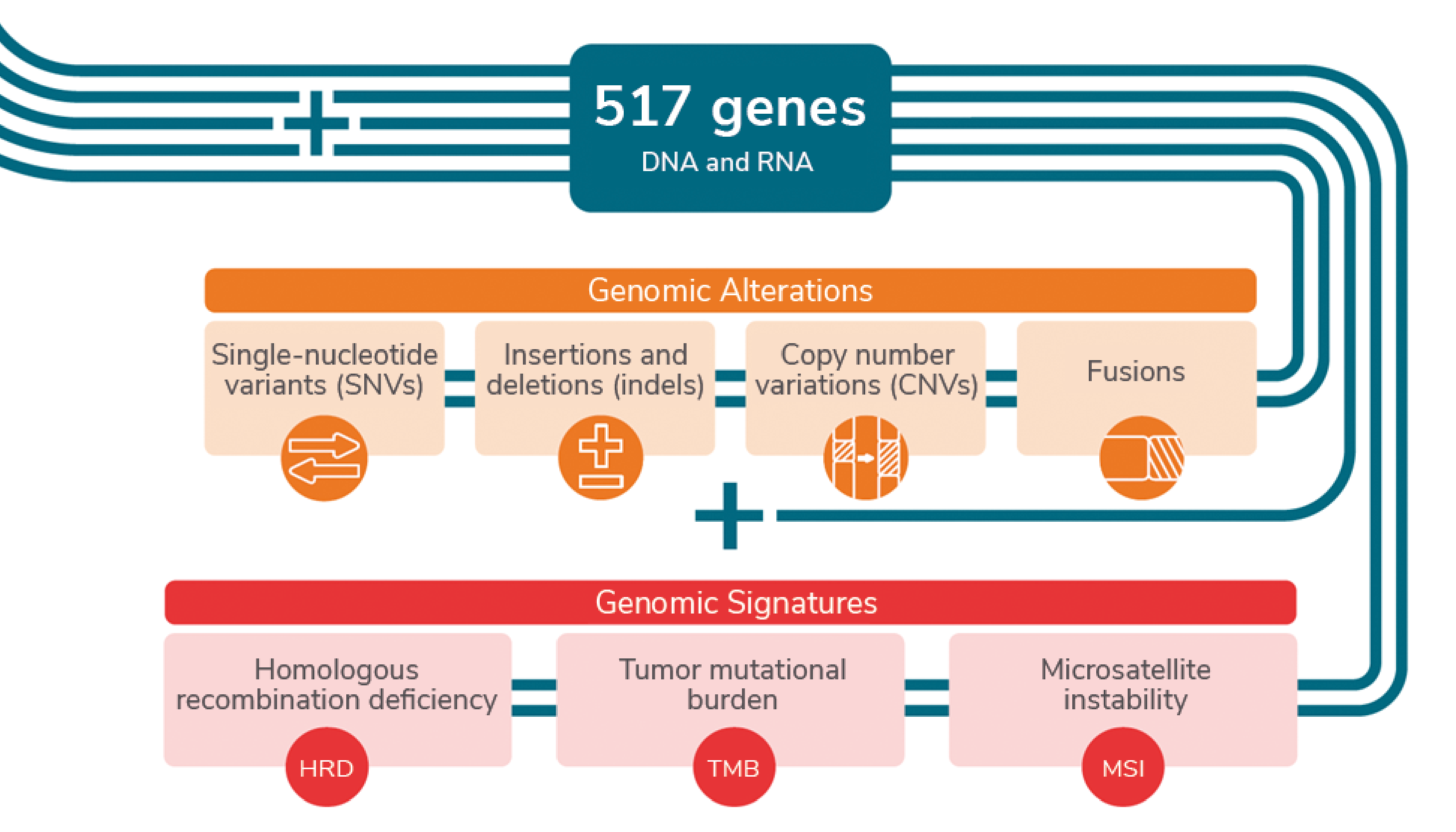
The Ion Torrent™ Oncomine™ Comprehensive Assay Plus, available on the Ion GeneStudio™ S5 System, is an amplicon-based CGP assay that brings the power of in-house CGP with a highly automated approach to meet the needs of labs at varying levels of NGS expertise. With highly automated library prep and sequencing systems that only require around one hour of hands-on time, the assay detects a broad range of genomic alterations – including single-nucleotide variants (SNVs), insertions and deletions (indels), copy number variations (CNVs), and fusions across 517 genes. Additionally, the assay detects complex biomarkers or genomic signatures such as homologous recombination deficiency (HRD), tumor mutational burden (TMB), and microsatellite instability (MSI).
Finding HRD causes and consequences
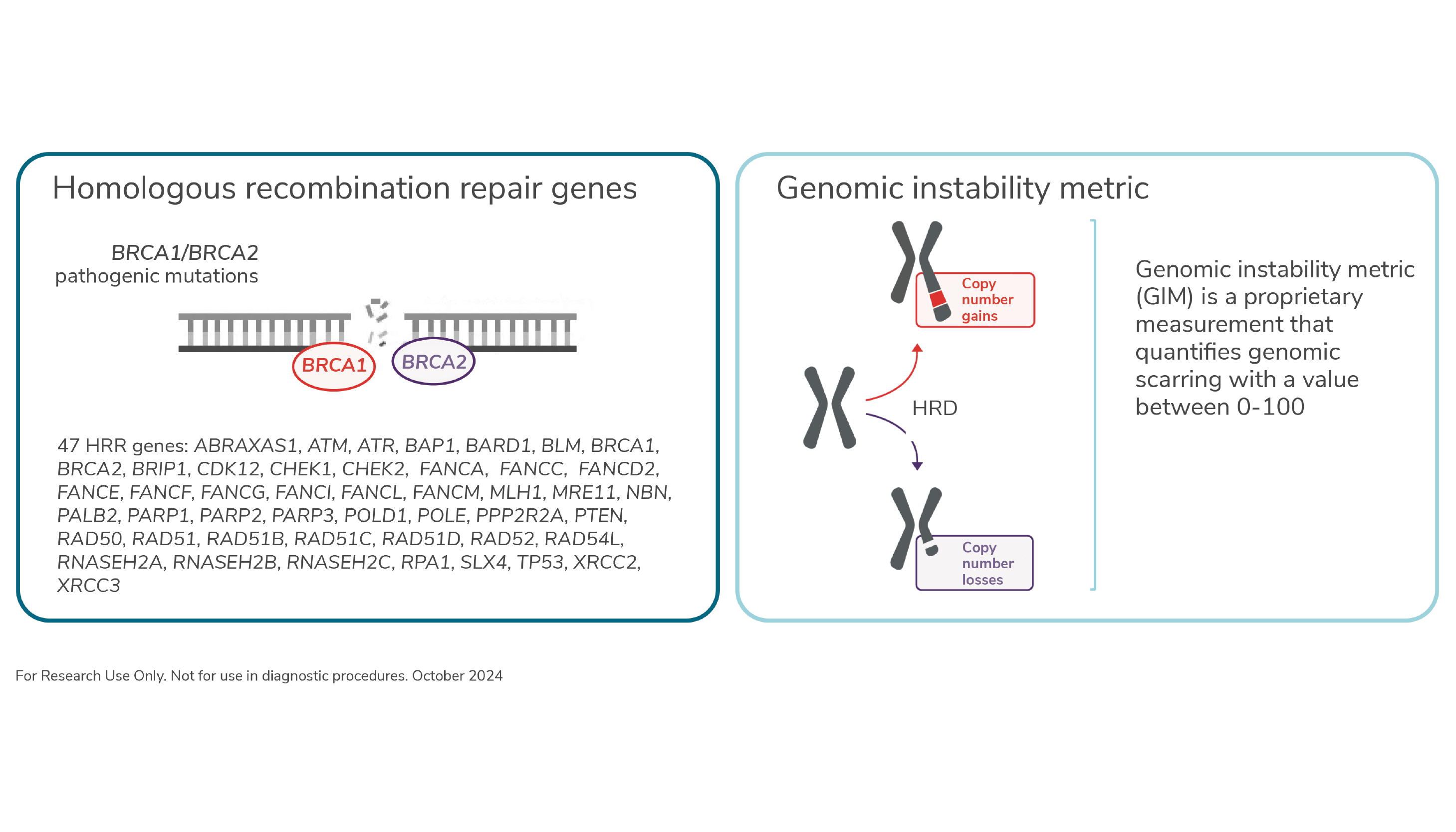
HRD is a phenotype that is characterized by the inability of a cell to effectively repair DNA double-stranded breaks using the homologous recombination repair (HRR) pathway (2). Of the genomic signatures, HRD is becoming increasingly relevant – especially in ovarian, breast, prostate, and pancreatic cancer – because of its association with poly (adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitors.
The Oncomine Comprehensive Assay Plus measures both the causes and consequences of HRD. Causes are assessed by detecting mutations in 47 genes associated with HRR, including large genomic rearrangements in BRCA1 and BRCA2. Consequences of HRD or genomic scarring are measured using a genomic instability metric (GIM) – a numeric value between 0 and 100 that summarizes the unbalanced copy number changes across the autosomes resulting from HRD with higher GIM values correlating with more genomic instability. Tumor research samples that have BRCA1/BRCA2 pathogenic mutations or markers of genomic instability are categorized as being HRD positive in ovarian cancer.
Case study: detecting HRD in ovarian cancer research samples
In a retrospective multicenter study of stage III–IV ovarian cancer research samples treated with chemotherapy from the MITO16/MaNGO-OV2 clinical study (n=100), HRD status was determined based on the presence of pathogenic mutations in BRCA1 and BRCA2 in combination with GIM using a predefined threshold of ≥16 to define high GIM (3). The Oncomine Comprehensive Assay Plus had good overall concordance with the reference method. Further studies will be needed to determine the appropriate thresholds for other cancer types, such as breast and prostate cancer.
Streamlining the workflow
When evaluating CGP assays, ease of use, turnaround times, and robustness with commonly encountered sample input requirements should all be considered. The Oncomine Comprehensive Assay Plus is a complete CGP solution that detects genomic alterations in 517 genes – plus genomic signatures such as HRD, TMB, and MSI – without the need for any additional add-on kits. One comprehensive assay for genomic profiling across tumor types greatly streamlines a lab’s workflow when it comes to operations, and logistics. In addition, as a single vendor of sample-to-report solutions, including instruments, consumables, analysis, and support, Thermo Fisher Scientific helps simplify the introduction of in-house CGP to your lab at varying levels of NGS expertise.
Learn more about the Oncomine Comprehensive Assay Plus at thermofisher.com/oncomine-ocaplus
For Research Use Only. Not for use in diagnostic procedures. © 2024 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified.
References
- E Jantus-Lewintre et al., “Multicentric evaluation of amplicon-based next-generation sequencing solution for local comprehensive molecular tumor profiling.” Poster presented at ESMO 2023; October 20-24; Madrid; Spain. Poster 219P.
- MD Stewart et al., “Homologous recombination deficiency: concepts, definitions, and assays,” Oncologist 27, 3 (2022). PMID: 35274707.
- N Normanno, “Future Clinical Perspective of HRD Testing in Ovarian Cancer Samples Using NGS CGP,” European Medical Journal Webinar (2023). Available at: https://www.emjreviews.com/type/webinars/





