Strength In Numbers
Sampling multiple tumor sites can reveal heterogeneity that would otherwise go unnoticed
At a Glance
- Sampling a single tumor site reveals what’s present at that location – but risks missing high-grade disease due to unevenly distributed heterogeneity
- Multi-site sampling can be accomplished affordably if multiple samples are processed in a single cassette
- This approach allows pathologists to look at several regions of the same tumor and greatly increases the odds of spotting areas of high-grade disease
- Sampling multiple sites requires a greater time commitment, but technology can help – and the added time may be worth it to optimize diagnosis and treatment
When you sample a tumor, what is it you’re really looking at? The answer may seem obvious, but what many people don’t realize is just how heterogeneous a single tumor may be. What may seem indolent at first glance may in fact turn out to be highly aggressive – but that’s something a pathologist might not discover by looking at only a few samples. Unfortunately, that’s how a lot of grossing is still conducted. So how can we change this to account for the level of heterogeneity that’s often present in cancers? I propose a new approach, which I’m pleased to say has now received good feedback by several internationally-recognized pathologists: multi-site tumor sampling.
A revealing review
The story started one day in 2012, when I went back to the grossing room to check whether or not a conventional low-grade clear cell renal cell carcinoma (CCRCC) had invaded the patient’s renal sinus. I confirmed in situ that it hadn’t – but as I was there anyway, I selected a few more samples for microscopic analysis. To my surprise, one of those samples showed some areas of high-grade carcinoma. I had to call the urologist and say, “Sorry; this isn’t a low-grade tumor anymore” – the sort of thing clinicians and surgeons refer to as “pathologists’ inconsistencies,” because they aren’t familiar with the complexity of our work.
But that case taught me to ask myself how much important information I was losing by following internationally accepted protocols for tumor sampling. And I did something that might have looked crazy to my colleagues: I began performing prospective total tumor sampling of CCRCCs, so that I could compare the information I obtained that way with what others got from standard methods. It was a risky decision – not for any medical reason, but because I was at high risk of being killed by my technicians due to the sudden increase in laboratory workflow. But after soliciting the help of some colleagues, I finally got 47 totally sampled CCRCCs… leaving me with over 1,400 slides to review!
At first, I examined only one slide per centimeter of tumor diameter, as per protocol – but later on, I began reviewing all of the slides. The results were very concerning: while routine sampling detected 7 high-grade tumors in 47 cases, my total sampling detected 17 (1). This means that, when using standard sampling protocols, pathologists may actually be giving incomplete information in their reports. Our methods need to change.
A dynamic disease
Why is this such a problem? Cancer is a dynamic process, and as it evolves, malignant cells develop different mutations in different areas of the tumor. This leads to “regionalization” – unique, stochastic and utterly unpredictable. We still don’t know the rules that govern this evolution. In the case of CCRCC, the molecular hallmark is the inactivation of the VHL gene, which codes for the von Hippel–Lindau tumor suppressor protein (pVHL). Aberrant pVHL provokes permanent intracellular activation of VEGF, leading to enhanced angiogenesis. Most modern CCRCC therapies are based on anti-angiogenesis – but because of intratumoral heterogeneity, some regions respond to treatment while others don’t. The ones that don’t are responsible for tumor progression.
Current sampling protocols were designed before we realized that heterogeneity was a major issue. Over the last few years, though, massive sequencing tools have shown us the full scope of the problem… but sampling protocols haven’t changed, and new problems require new solutions. The first step in this solution is thorough tumor sampling to learn as much as we can about each neoplasm’s individual peculiarities. Because total sampling is impossible in many tumors due to their size, we must choose which parts to analyze – and being as thorough as possible is our responsibility as pathologists. That’s where the promise of the multi-site tumor sampling strategy comes in.
From my own experience (that is, the crazy study I conducted in 2012), I knew that “the more you sample, the more you find.” The question is where to stop. How do you balance finding heterogeneity and establishing a sustainable system? The only way I could envision affordably sampling more tumor regions was to take a higher number of smaller fragments and put several of them on the same cassette for analysis – keeping the total number of cassettes the same, but increasing the number of samples we could examine for the same price. That’s what I call the multi-site tumor sampling (MSTS) protocol. The catch? This was all based purely on common sense; I didn’t have any objective data to prove that several small fragments from distant tumor regions were more informative than a single large fragment. What I needed to do next was cooperate with a scientist – Jesús Cortés, a physicist who specializes in modeling and data mining (see box, “The Physicist’s Story”, below). Together, we were able to demonstrate that MSTS outperforms routine sampling, and that it does so without incurring extra costs (Figure 1) (2)(3)(4).
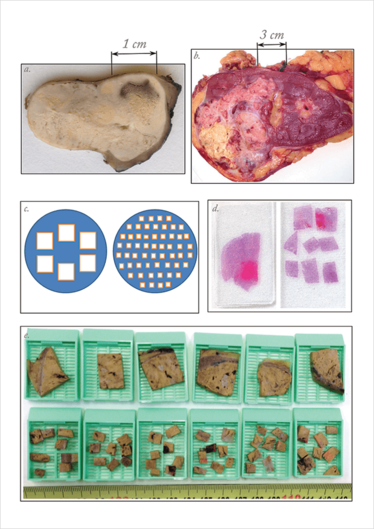
Figure 1. ITH in CCRCCs may be hidden (a) or evident (b) to the naked eye during the management of surgical specimens, an issue that is critical for subsequent tumor sampling. The RS strategy selects for analysis 1 sample per cm of tumor diameter, as reflected at the left side of panels c–d (c, diagram d, histological slide) and at the top row of blocks in panel e. In contrast, the DAC strategy selects more small-pieces for tumor sampling (8 pieces in this example, versus 1 for RS) but the pieces are randomly chosen along the tumor. Importantly, both RS and DAC methods demand the same laboratory costs. ITH, intratumor heterogeneity; CCRCC, clear cell renal cell carcinoma; RS, routine sampling; DAC, divide-and-conquer. Adapted from (3).
The Physicist’s Story
By Jesús M. Cortés
In the “ideal” situation, where heterogeneity is randomly distributed across the tumor, sampling one large piece of tumor is equivalent to sampling many small pieces. But that’s not how it works in real life. Heterogeneity is regionally, not randomly, distributed – and that means the two strategies have obvious differences with very real consequences for patients. To prove it, I used a simple modeling approach to show the clear advantages of one strategy (namely MSTS) with respect to the other (routine sampling, or RS) in detecting intratumoral heterogeneity. The approach we used is the “divide and conquer” algorithm (3), a well-known strategy for solving practical problems in computer science. Briefly, it involves dividing a complex problem into smaller ones that can be solved independently and merged into an overall solution. In this case, that meant revealing the superiority of MSTS as a sampling technique for intratumoral heterogeneity detection.
My main research interests lie in systems neuroscience and neuroimaging – but alongside that work, I will continue to do my best to put experts’ intuitions into numbers, just as I’ve done in this project. Why do I do it? Because I am truly convinced that clinical research can benefit from the methods already well-established in the quantitative sciences.
Jesús M. Cortés is Ikerbasque Senior Researcher and Head of the Quantitative Biomedicine Group at Biocruces Research Institute, Barakaldo, Bizkaia, Spain.
The start of a sampling revolution?
MSTS isn’t limited to CCRCCs, though; it can be applied to any tumor that can’t be sampled in its entirety, and it’s especially advisable for neoplasms known to have high intratumoral heterogeneity. We’ve shown that MSTS is much more effective than routine sampling in detecting intratumoral heterogeneity, and it can easily be combined with molecular testing – but there’s a downside: it’s also much more laborious for pathologists in the grossing room. That could be a major obstacle to widespread implementation. To overcome this hurdle, we recently proposed the use of a cutting device that significantly reduces sampling time (2). It’s important to pave the way for MSTS as much as possible, because I truly believe it’s the method we need to use with large tumors to meet oncologists’ expectations.
We have already demonstrated in silico that MSTS is more efficient than routine sampling in detecting intratumoral heterogeneity (3). Now we have finished the first clinical validation of our protocol, based on the evaluation of classic histopathological parameters – and the results confirm our expectations. In fact, MSTS detected a significantly higher number of tumors with high-grade areas in a series of 38 CCRCCs, even though the speed, quality and cost of the two techniques are comparable (Table 1) (4). Right now, we’re developing more clinical validations of the method, so we’re looking forward to even more interesting data by the end of the year.
| Histological parameters | MSTS | RS | P value (X2 test) |
| High grade (G3/4) | 31 | 21 | 0.0136 |
| Granular eosinophilic cells | 32 | 22 | 0.0114 |
| Sarcomatoid phenotype | 12 | 6 | 0.1 |
| Tumor necrosis | 10 | 7 | 0.5 |
Table 1. Comparison between both sampling protocols showing that MSTS outperforms RS. MSTS, multi-site tumor sampling; RS, routine sampling (4).
Adoption couldn’t be simpler
We’re now using MSTS routinely in our own laboratory, because it has proven to be so advantageous in large tumors. How can other labs adopt our method? It’s fairly straightforward: when making the paraffin block, simply put six to eight tissue fragments in the same block instead of only one – not a big change, and the only difference between routine sampling and MSTS! I hope that the method’s simplicity and its benefits will convince other pathologists to consider it, but I know that will take some time.
My advice to others is to seriously consider whether or not you’re really optimizing your own work. If you aren’t, make the changes needed to benefit your patients as much as possible. I think we have developed somewhat of a contradictory attitude – making microscopic and molecular studies our most important goal, considering the macroscopic analysis of tumors a secondary task, and leaving fundamental work like tumor sampling to the residents. We need to return our attention to those aspects of our work, and to update our most basic protocols. Keep in mind that the success of our expensive and sophisticated devices depends on a single, humble decision: how much tumor must I sample, and in what way, to efficiently detect intratumoral heterogeneity?
José I. López is Head and Professor of Pathology at Cruces University Hospital, University of the Basque Country, and Senior Researcher at Biocruces Research Institute, Barakaldo, Bizkaia, Spain.
- JI López et al., “Grade heterogeneity in clear ell renal cell carcinoma”, BJU Int (2013).
- JI López, JM Cortés, “A multi-site cutting device implements efficiently the divide-and-conquer strategy in tumor sampling”, F1000Res, 5, 1587 (2016). PMID: 27540472.
- JI López, JM Cortés, “A divide-and-conquer strategy in tumor sampling enhances detection of intratumoral heterogeneity in pathology routine: A modeling approach in clear cell renal cell carcinoma”, F1000Res, 5, 385 (2016). PMID: 27127618.
- R Guarch et al., “Multi-site tumor sampling (MSTS) improves the performance of histological detection of intratumoral heterogeneity in clear cell renal cell carcinoma (CCRCC)”, F1000Res, 5, 2020 (2016). PMID: 27635226.
José I. López is Head and Professor of Pathology at Cruces University Hospital, University of the Basque Country, and Senior Researcher at Biocruces Research Institute, Barakaldo, Bizkaia, Spain.