Cancer’s Circulating Secrets
Liquid biopsy is a powerful research tool that allows investigators to better understand cancer’s intricacies without subjecting patients to invasive procedures
sponsored by Thermo Fisher Scientific
Liquid biopsy is an ever more vital component of cancer research. Circulating tumor DNA (ctDNA) offers a window into the characteristics, the changes, and the vulnerabilities of tumors, and as a result, its adoption in research laboratories is becoming more widespread. Alberto Bardelli, an expert in cancer genetics and personalized medicine, explains how liquid biopsy has benefited his work – and why he thinks more researchers should take advantage of it.
How did you begin using liquid biopsy in your research?
I first became interested in the technique because of my work on cancer clonal evolution and drug resistance. A number of years ago, my colleagues and I reported that colorectal cancer patients who respond to EGFR blockade therapy do so for a limited period of time, then relapse (1). We needed a way to identify the mechanisms of secondary drug resistance and patients who were likely to relapse – and the best way of doing so at the time was to take tissue biopsies. It was difficult to engage patients in the protocol, though, because it was invasive and didn’t provide them with an immediate, tangible benefit. We found ourselves running in circles with no way to identify the cancer’s mechanism of resistance to this targeted therapy.
Then I thought, “Perhaps we can use blood as a surrogate for tissue.” There was already evidence that ctDNA was a potential source of information for this type of research (2) – and so, starting in 2009, we began to build our ctDNA collection. Ever since, in every clinical trial we have designed, we have collected blood and isolated plasma and peripheral blood mononuclear cells (PBMCs) from our patients. Thanks to those ctDNA samples, we were able to uncover the mechanism of EGFR blockade resistance in colorectal cancer patients (1). And that made us realize that liquid biopsy could be an incredible source of information on how tumors evolve during therapy.
A few years later, we asked the question, “When treatment stops, will resistant clones in the blood increase, decrease or remain constant?” To our surprise, we found that, when therapy was suspended, the resistance mutations we saw in ctDNA would drop almost immediately (3). We’re now beginning a clinical trial called CHRONOS – in which, for the first time in colorectal cancers, clinicians will make treatment decisions based on liquid biopsy. It will be a large trial (>250 patients), and our plan is to track levels of KRAS mutation in the blood during anti EGFR therapy and use those to determine when to restart the therapy.
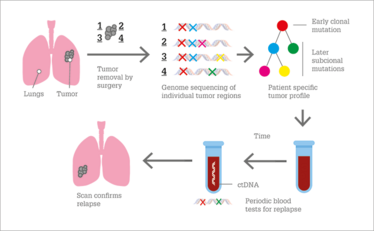
Figure 1. The use of liquid biopsy to monitor for cancer relapse after surgical excision. Adapted from (5).
How can liquid biopsy benefit cancer research?
It can answer key research questions. For instance, which cells release ctDNA most effectively? If you have a patient with metastatic colorectal cancer, will the ctDNA recapitulate the disease progression? Will it accurately represent all the lesions present in that patient?
One way to address this is by coupling liquid biopsy to “warm autopsy” – the practice of procuring samples soon after death. We have designed a rapid donation trial (DONUM) to collect tissue and blood from patients with multi-organ disease to understand how each tumor contributes to the total ctDNA pool. Eventually, we hope to understand enough about individual tumor contributions to study patients with metastatic disease primarily through their ctDNA. Ultimately, I hope the knowledge we gain will lead to better disease tracking over time, better monitoring of minimal residual disease, and – one day – large-scale screening for early cancer detection.
Five years ago, you would go to a meeting and find perhaps one or two presentations on ctDNA studies. Now, entire sessions are focused on liquid biopsy; there’s been incredible interest. We now need to define clinical settings in which liquid biopsy can inform treatment approaches. We have already shown it’s a valuable approach to track tumor dynamics and identify mechanisms of drug resistance, in the future it might be used to prescribe drugs, determine the need and timing of CT scans and other tests, assess minimal residual disease, decide when further chemotherapy is warranted, and when it is not…
How does research benefit from NGS and biomarker analysis?
Next generation sequencing (NGS) and multi-biomarker panels are crucial to understanding and improving therapies that specifically target tumor cells. In colorectal cancers, we discovered the importance of KRAS, then BRAF, then NRAS… Now we’re looking at alterations like HER2 and MET amplifications and MAP kinase mutations. The list is constantly growing – who knows what we may need next? Cancer is in essence a genetic disease, so next-generation sequencing of DNA and RNA will be even more relevant in the future than it already is.
The more we know about cancer, the more we realize that it continually changes – and especially so during treatment. Both kinase-targeted therapy and immuno-oncology will benefit tremendously from NGS approaches – and, in those fields, the research of today will yield the diagnostics of tomorrow. In the future, we will want to predict how many and which neoantigens are present in a tumor at a given moment, we already need to quantify the number of mutations per megabase in individual tumors. This information comes from NGS analyses, so this technology is poised to make a huge impact. We particularly need two things: platforms that simplify the process so that users don’t need a dedicated bioinformatics team to understand their results, and standardization so that laboratories can work with one another’s data.
What do you expect from the field in the near future?
I can think of a few advances that would be especially useful. Panels that are capable of tracking tumor evolution, for instance, can help determine when a patient may need to change treatments and what the next option should be. NGS approaches to identify neoantigens will be valuable. So too will be knowledge of T cell receptor rearrangements to study the immune system’s reaction to a tumor. Analyses like these are expensive, but within reach – recently, for the first time, a personalized approach was prospectively used to track individual lesions that evolved in lung cancer patients (4). Perhaps in the future, every patient will be followed with an individualized multi-biomarker panel. And eventually, as the cost of sequencing decreases and the sensitivity of liquid biopsy improves, perhaps tumors will be interrogated longitudinally in routine clinical practice – using ctDNA as a source of information.
- S Misale et al., “Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer”, Nature, 486, 532–536 (2012). PMID: 22722830.
- F Diehl et al., “Circulating mutant DNA to assess tumor dynamics”, Nat Med, 14, 985–990 (2008). PMID: 18670422.
- G Siravegna et al., “Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients”, Nat Med, 21, 795–801 (2015). PMID: 26030179.
- C Abbosh et al., “Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution”, Nature, 545, 446–451 (2017). PMID: 28445469.
- A Bardelli, “Personalized test tracks cancer relapse”, Nature, 545, 417–418 (2017). PMID: 28541318.
Alberto Bardelli is Professor of the Department of Oncology, University of Torino, Director of the Laboratory of Molecular Oncology at the Candiolo Cancer Institute-IRCCS, Torino, Italy, and President Elect of the European Association for Cancer Research.