Color Truth for Infectious Disease Imaging
Color is vital in a field where special stains are key to diagnosis
Color management can enhance whole-slide imaging in infectious disease diagnosis. Color is a key aspect of accurate diagnosis, which is critical to effective treatment. There is a drive to automate diagnosis and improve accuracy – but this is hindered without the ability to standardize digital devices to real-world colors.
When Hans Christian Gram first discovered that bacterial species could be visually differentiated by the binding of crystal violet to peptidoglycan cell walls, he set in motion a methodology that would revolutionize infectious disease diagnosis and analysis.
Pathogenic species of fungi, protozoa, and bacteria can be thought of as “alien” cells on a background of expected human cells. These pathogens can be discerned through stains that bind infectious agents in a visibly different way to the patient’s native tissue – creating a rapid, universal toolkit for diagnosing diseases and matching symptoms to their causative organisms. Even today, color is central to a diagnostician’s daily work.
Then and now
In the early days of antibiotics, researchers quickly discovered that beta-lactam antibiotics, such as penicillin, would only kill bacteria with high cell-wall peptidoglycan content. As a result, the colorful output from a positive Gram stain became a routine indication for treating infections with penicillin.
In modern-day medicine, the scope of infectious and communicable disease has grown exponentially, reciprocated by a wealth of colorful histopathology staining techniques to aid an ever more complex diagnostic process. This means much greater differentiation of increasingly specific infectious diseases can be made during histopathological diagnoses, with each colored stain response acting as a unique identifier on which patient treatment decisions can be made.
Digital pathology – and, in particular, whole-slide imaging (WSI) technology – may overcome caveats to the sharing and analysis of data for a greater breadth of diagnoses in a growing population. Obviously, bright-field light microscopy has some limitations with respect to infectious disease; it cannot resolve most viruses, or even the smallest bacteria, without oil immersion. Nonetheless, some of the world’s leading WSI OEM developers and vendors, supported by the brightest optical engineering minds on the planet, are developing technologies that can confidently visualize more species of bacteria, fungi, and protozoa at the limits of bright-field resolution. Although most WSI systems are designed for tissue pathology (and thus limited by resolution and lack of oil immersion), they are increasingly used for infectious disease diagnoses. With further exploration into this novel use of WSI, we can increase the adaptability, ubiquity, and potential of digital pathology devices and analytical software (1). Armed with a catalogue of colored special stains and resolution limits at the cutting edge of modern WSI optics, we can identify some of the world’s most common and clinically relevant infectious diseases (see Figure 1).
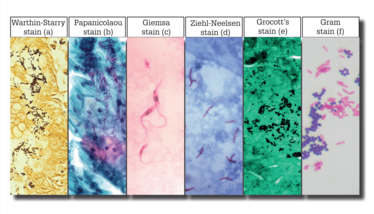
Figure 1. a) Helicobacter pylori and spirochete species; b) Candida fungal species; c) Trypanosoma cruzi; d) Mycobacterium species; e) Histoplasma fungal species; f) generic bacterial species, further identifiable by their cellular structures and biochemistry.
The benefits are not limited to human diagnostics – veterinary and plant science play a significant role in ensuring agricultural supply, development of new antibiotics in the pharmaceutical industry, and furthering the understanding of disease persistence and antibiotic resistance in academic and industrial study. The ability of modern WSI scanners to optically resolve these disease agents leads to an interesting new avenue for clinics and researchers – and, like all diagnostic pathways, ensuring the standardization of data reliability, fidelity, and quality across multiple sources forms part of confidence in deployment.
Color cues
Because the colorful special staining of many of these infections is a key differentiator against the host tissue or liquid biopsy background, it is vital to ensure that the digital colors in WSI images remain faithful to their intended targets. Only then can they provide human or artificial intelligence (AI) analysts with the most valid information from which to make diagnostic decisions. The digitization process, despite offering great advances in data capture rate, storage, and automated analysis, also carries with it some inherent challenges – including accurate color reproduction. Without careful management, this intrinsic caveat of digital imagery could misrepresent the colors used as diagnostic indicators of infectious diseases.
WSI systems often contain basic color handling functions in their image files – from rudimentary, idealized lookup tables to inbuilt physical targets that update color responses on the fly. Unfortunately, many of these functions are limited or biased by their intended applications in clinical oncology and pharmaceutical development and may be in formats that are not representative of the real spectral responses of stained samples. For a WSI system to be universally applicable to infectious disease requires an external, unbiased method that can evaluate the color output from any system. This can identify inherent errors that differ across WSI vendors and provide a mechanism for not only standardizing the color output of these systems, but also ensuring that onward analysis is based on the real color in the original stained sample.
One solution is to apply device-specific International Color Consortium (ICC)-standardized color profiles to all WSI-generated images using real-world spectral data found in stained infectious disease samples. This can be achieved by using a slide with histologically stained patches that precisely mimic real-world colors – for histopathology, common stained tissues; for infectious disease analysis, special stains to identify pathogens. By measuring the spectral absorption of commonly used stains on a tissue-mimicking slide and comparing those measurements with an image of the same slide as scanned by the pathologist, the interpreted error (the scanner’s deviation from a standard truth) can be calculated. ICC profile metadata is then generated and used to correct the pre- or post-imaging output so that the digital image contains the ground truth colors.
The color is therefore not only closer to truth, but also unified across all scanners. For human analysis, this means that diagnosis is based upon the same data as in the real sample. In a future increasingly moving toward big data and AI software, applying calibration technology on a regular basis ensures that decisions are based on uniform input – and that all images, regardless of source, remain digitally unaltered and are quality-controlled to be ground truth color as standard. The power that such technology represents can be seen in the ability to correct large errors and high variation across many WSI systems in the spectral response ranges commonly used in infectious disease staining (see Figure 2).
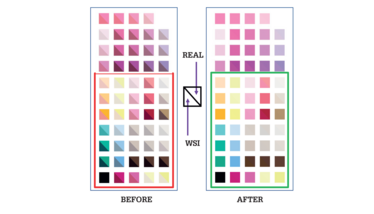
Figure 2. An effective slide-based color calibration slide must cover the whole histopathological spectrum to ensure the optimum coverage of special stains used in infectious disease diagnosis (boxed region - bottom), as well as the common H&E range (unboxed region - top). The inherent errors of all WSI systems (“Before”) can be corrected to the real color in the special-stained images (“After”) through the use of an ICC profile calculated from measurements of slide patches that accurately represent ground truth color data.
WSI systems must be at peak reliability and data quality to image and detect infectious agents at the current extremes of the technology’s reach. For that to happen, we must have control over any potential imaging artifacts, such as color misrepresentation, that could skew the sensitive analysis. As for the motivations for general clinical WSI color calibration, the 2016 FDA guidance suggests, “The WSI system should be tested with a target slide. The target slide should contain a set of measurable and representative color patches, which should have similar spectral characteristics to stained tissue” to achieve this level of confidence, fidelity, and validity (2). With big pharma equally concerned with achieving peak levels of GLP acceptability for drug pipelines (such as those for developing new antibiotics) and software analysis vendors looking to provide solutions that encapsulate and facilitate this drive for standardization and QA, there is a reason for appropriate image data color management in every sector of the WSI market.
The market for WSI systems and data analysis is growing rapidly – which means vendors are under more pressure than ever to be both competitive and in line with emerging accreditation. The field of infectious disease diagnosis is rich with successful technologies – RT-PCR, next-generation sequencing, an array of molecular tests for liquid biopsies – that WSI must match or exceed in throughput and fidelity. The boundaries of imaging speed and quality are likely to be pushed even further, increasing the range of infectious disease detection and therefore increasing the need for stringent data regulation in more clinical applications. Meeting regulatory guidelines is paramount to success, and slide-based color calibration technology is well-positioned to not only provide accuracy and reliability for the current state of play, but also future-proof WSI technologies for use in infectious disease diagnostics.
- DD Rhoads et al., “Comparison of the diagnostic utility of digital pathology systems for telemicrobiology”, J Pathol Inform, 7, 10 (2016). PMID: 27076988.
- US Food and Drug Administration, “Technical performance assessment of digital pathology whole slide imaging devices” (2016). Available at: bit.ly/2WERBH5.
Richard Salmon is Product Manager for Life Sciences at FFEI, Hemel Hempstead, UK.