Giving Cancer the Brush-Off
WATS3D – a new brush-based biopsy technique combined with computer-assisted analysis – can improve detection rates for pre-cancerous conditions in patients with Barett’s esophagus
At a Glance
- Forceps biopsy screening for esophageal pre-cancers has a high degree of sampling error
- Inter-observer variability further limits the protocol’s ability to detect significant disease
- A new technique, WATS3D, combines wide-area trans-epithelial sampling with 3D computer-assisted analysis
- WATS3D reduces sampling error, improves detection of goblet cells and dysplasia with assistance from computer-generated images, and highlights the most atypical areas of tissue
Esophageal cancer may not be among the most common forms of cancer, but it’s certainly one of the most fatal. Only about one in five people diagnosed with esophageal cancer will survive five years (1), and over two-thirds are diagnosed with late-stage disease (2). The key precursor to esophageal cancer is Barrett’s esophagus (BE), a metaplastic change that is often followed by transition to dysplasia. Patients who exhibit signs of BE or esophageal dysplasia (ED) must undergo increased surveillance or ablation so that cancer can be preempted. However, current screening and surveillance technologies are flawed – a dangerous situation for patients at risk for esophageal cancer.
What’s the current gold standard for identifying BE and ED?
The current gold standard for screening and surveillance of BE and associated neoplastic lesions is forceps biopsy of patients with known or suspected BE. The Seattle protocol requires biopsies from all four quadrants of Barrett’s mucosa – every centimeter for patients who have dysplasia; every two centimeters for those who don’t – as well as any visible lesions. The diagnosis is based on histologic evaluation of mucosal biopsies.
However, this method is fraught with problems. Foremost of these is that it results in a high degree of sampling error because the biopsies cover only a small fraction of the Barrett’s mucosa (in some cases, less than 5 percent). Several studies have shown that up to one-third of BE patients present with high-grade lesions and even cancer within one year of endoscopic screening or surveillance (3). This indicates that sampling error resulting in false negatives is a huge problem with the Seattle protocol.
The second problem is inter-observer variability regarding establishing a diagnosis of dysplasia in forceps biopsies. Although pathologists have been assessing biopsies for several decades now, we haven’t seen any significant gain in inter-observer agreement, whether general or expert, over that period of time. Pathologists still differ quite strongly in interpretation of dysplastic lesions, particularly when it concerns distinguishing regenerating epithelium from low-grade dysplasia – a common problem – and secondarily differentiating low-grade from high-grade dysplasia. These are critical decisions because non-dysplastic, low-grade dysplastic, and high-grade dysplastic BE may be treated differently, so an accurate diagnosis is important for patient care.
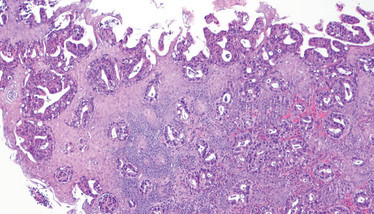
Adenocarcinoma of the esophagus. Credit: Wikimedia user CoRus13.
Tell us about the wide sampling approach…
It’s called wide-area transepithelial sampling with 3D computer-assisted analysis (WATS3D). From the name, you can probably determine that the test consists of two major components aimed at decreasing the common problems associated with the Seattle biopsy protocol.
WATS3D uses a proprietary brush composed of longer and harder needles than conventional soft-brush cytology brushes. Soft brushes typically acquire superficial exfoliated cells from the top portion of the mucosa, but dysplasia and early cancer often reside in the deep mucosal crypts. If one doesn’t want to miss those areas, one needs to acquire tissue from deeper in the mucosa. The WATS3D brush is useful because it acquires full-thickness mucosal fragments – that is, tissue all the way down to the muscularis mucosa. And because it’s a brush that sweeps across the mucosal surface and acquires a lot of tissue, it greatly decreases sampling error. We’ve seen significant increases in detection rates for both goblet cells (required for fulfillment of the BE criteria) and dysplasia using the WATS3D diagnostic platform.
The second aspect of WATS3D is computer-assisted analysis. The computer system scans all of the tissue acquired by the brush, then uses a neural network and artificial intelligence (AI) to analyze it. This analysis includes a battery of morphologic assessments on the nuclear characteristics of the tissue; it ranks tissue fragments according to degree of atypicality; and it performs a unique extended depth-of-field (EDF) imaging analysis on the cell aggregates. Ultimately, it synthesizes images of the most atypical tissue fragments and presents them to the pathologist on a computer screen to be used when the pathologist signs out the specimen at the microscope.
In EDF imaging, tissue fragments – typically between 150 and 200 microns thick on a WATS3D smear – are scanned at 5 μm intervals all the way through the tissue, similar to a CT scan of the human body. The system then uses neural network programming to synthesize all of those 3D cuts into one crisp, clear, artifact-free 3D image. That way, the pathologist has a unique view of the tissue fragments, including whole crypts and their features, which leads to improvement in diagnosis. In one recent study, the degree of inter-observer agreement was much higher for WATS3D specimens (all kappa values >0.7) than for forceps biopsies alone (4).
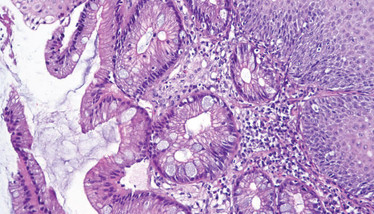
Biopsy from the tubular esophagus showing incomplete intestinal metaplasia. Credit: Wikimedia user CoRus13.
Tell us about your own experience with WATS3D…
WATS3D specimens come in two forms. The first is a formalin-fixed, paraffin-embedded, H&E-stained slide similar to a normal mucosal biopsy. However, the fragments of tissue are very small – I would consider them “micro”-biopsies. The other specimen is the tissue smear I’ve described above. I refer to these two approaches as “histocytology,” because we can see clear histologic features of dysplasia in what amounts to cytology brush specimens. The addition of the computer analysis is unique – and it’s fascinating, because you get to see crypts almost in their natural form.
Overall, the learning curve for WATS3D is very short. I have a lot of experience diagnosing BE via traditional means – and, after I went through a teaching process of about 20 cases, I felt very capable of diagnosing the vast majority of WATS3D cases. By the time I hit 30 cases, I felt extremely comfortable. Despite the novel technique, one evaluates all the same features in WATS3D specimens that one sees in traditional forceps biopsies, so it’s really nothing new from the histologic point of view. I view it as a crisper, more visually accurate and specific method of evaluating tissue.
The recommendation right now is to use WATS3D with any patient who is endoscopically identified as having salmon-colored mucosa in the esophagus and therefore suspected or previously known to have BE. Why? Because WATS3D is proven to be more efficacious for both identifying goblet cells (that can help confirm the diagnosis of BE) and for identifying neoplastic precursor lesions. In my opinion, the technique should be used not only for surveillance, but also during each patient’s initial index endoscopy (screening), because most dysplasia and early cancer that we see in BE is detected at that point in time (prevalent dysplasia). That’s when the chances of finding dysplasia are greatest, so that’s when we should be using our most powerful detection tool.
Why is it so important to detect these conditions early?
At the moment, 95 percent of esophageal cancers are diagnosed outside the setting of surveillance (5). Even among patients in a surveillance program, at least one-third develop cancer within a year of an endoscopic surveillance (3). It’s clear that endoscopic surveillance in its current form is not efficient. We recognize from our experience with cancers in other parts of the body, such as in the cervix and the colon, that early (often endoscopic) detection of neoplastic precursor lesions can drastically reduce the incidence of invasive cancer worldwide. The same is true for esophageal cancer, which is particularly deadly.
To make headway in helping patients with this condition, we must work to detect its precursors and prevent its development, because once a patient has esophageal cancer, the fatality rate is high. We know that cancer develops through a metaplasia–dysplasia–cancer pathway. We know that when a patient transitions from metaplasia to dysplasia, regardless of how small or isolated the lesions, they have already acquired the necessary mutations to be considered at high risk for progression to cancer. There’s no reason we wouldn’t want to eliminate it at that stage, so it makes biological and ethical sense to try to find and eliminate pre-cancer before it develops into invasive cancer.

What are the next steps for WATS3D?
WATS3D is ready for routine clinical use. There have been five prospective trials (some randomized) and an inter-observer study that have all shown a greatly increased detection rate of BE and ED by WATS3D when used as an adjunct to mucosal biopsies. Those studies have been performed in both high-risk academic centers and low-risk general community populations with uniformly positive results. As a result, I believe that if you or I had BE, we would be better off having forceps biopsies plus WATS3D, rather than forceps biopsies alone, because the data unequivocally show that this combination approach has a greater chance of detecting pre-cancer, which is the goal of surveillance. Now that we have a safe and effective ablation treatment for pre-cancer (which was formerly treated by esophagectomy), WATS3D is ready for prime time.
Of course, our knowledge will continue to grow. There are studies currently underway to evaluate WATS3D in various patient groups, such as post-ablation, short-segment BE, low- and high-risk European and North American patients – so we’ll have even more information in a few years’ time than we do now. The field of BE is also moving toward biomarker-based evaluation of patients with non-dysplastic BE to determine which patients are at risk for progression and which aren’t, so that we can modify our existing surveillance programs accordingly. So far, randomized, long-term outcome studies in patients without dysplasia haven’t revealed any efficacious non-histological biomarkers, so that’s where I think we’re headed next. We need the ability to predict which patients without neoplastic precursor lesions are at risk of developing them. Luckily, because WATS3D tissue fragments are virtually all epithelial (with very little lamina propria and soft tissue), they are primed for molecular biomarker studies. I think, in the future, we’ll see WATS3D brush specimens used not only for morphological evaluation, but also in concert with biomarker panels.
Regardless of where the next few years take us, I think the data are already so compelling that WATS3D should already be considered standard of care – and I hope the rest of the world soon agrees.
Goblet Cells
An ongoing controversy in the definition of Barrett’s esophagus
In the US, the major governing bodies in gastroenterology all require two criteria to establish the diagnosis of BE. One is endoscopic recognition of salmon-colored mucosa in the esophagus, for the most part greater than 1 cm in length; the second is confirmation of the presence of goblet cells in biopsies from those areas of salmon-colored mucosa. In many other parts of the world, BE can be diagnosed by endoscopic evidence of salmon-colored mucosa alone (in other words, those governing bodies do not require pathologic identification of goblet cells to establish a diagnosis).
So why were goblet cells incorporated into the diagnostic criteria in the US? It’s partly based on the belief that only patients who have goblet cells in esophageal mucosa are at risk of neoplastic progression. However, we now have plenty of data to suggest otherwise. Many patients can develop dysplasia and cancer in esophageal columnar mucosa without goblet cells – something that has been proven in scientific cloning and lineage experiments (6).
It’s true, though, that most patients who do develop dysplasia and cancer do have goblet cells – even though they aren’t strictly required. And most gastroenterologists who see endoscopic evidence of salmon-colored mucosa in the esophagus, even if the biopsies fail to reveal goblet cells, still put those patients into a surveillance program, because there’s always a risk that the lack of goblet cell identification was due to sampling error. The sensitivity and specificity of biopsies to detect goblet cells, particularly in patients with shorter segments of BE, is very low.
- ASCO, “Esophageal cancer: statistics” (2019). Available at: bit.ly/2JjlKpU. Accessed May 13, 2019.
- Cancer Research UK, “Oesophageal cancer incidence” (2014). Available at: bit.ly/2JgmKeh. Accessed May 13, 2019.
- K Visrodia et al., “Magnitude of missed esophageal adenocarcinoma after Barrett’s esophagus diagnosis: a systematic review and meta-analysis”, Gastroenterology, 150, 599 (2016). PMID: 26619962.
- PR Vennalaganti et al., “Inter-observer agreement among pathologists using wide-area transepithelial sampling with computer-assisted analysis in patients with Barrett’s esophagus”, Am J Gastroenterol, 110, 1257 (2015). PMID: 25916227.
- NJ Shaheen et al., “ACG clinical guideline: diagnosis and management of Barrett’s esophagus”, Am J Gastroenterol, 111, 30 (2016). PMID: 26526079.
- R Odze, “Histology of Barrett’s metaplasia: do goblet cells matter?”, Dig Dis Sci, 63, 2042 (2018). PMID: 29998421.
Professor of Pathology at Harvard Medical School and Chief of Gastrointestinal Pathology at Brigham and Women’s Hospital, Boston, USA.